8AYJ
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiens complexed with 3-aminooxypropionic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
1U3A
 
 | | mutant DsbA | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Thiol: disulfide interchange protein dsbA | | Authors: | Serre, L. | | Deposit date: | 2004-07-21 | | Release date: | 2005-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intriguing conformation changes associated with the trans/cis isomerization of a prolyl residue in the active site of the DsbA C33A mutant
J.Mol.Biol., 347, 2005
|
|
8BBM
 
 | |
1U3J
 
 | | Crystal structure of MLAV mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
8AXP
 
 | |
1U3P
 
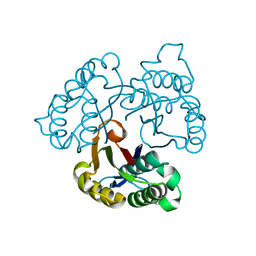 | | IspF native | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ZINC ION | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hecht, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
8B7S
 
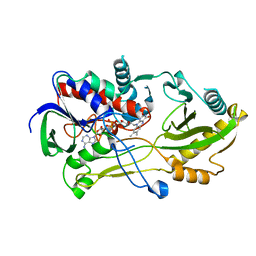 | | Crystal structure of the Chloramphenicol-inactivating oxidoreductase from Novosphingobium sp | | Descriptor: | Chloramphenicol-inactivating oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, L, Toplak, M, Saleem-Batcha, R, Hoeing, L, Jakob, R.P, Jehmlich, N, von Bergen, M, Maier, T, Teufel, R. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial Dehydrogenases Facilitate Oxidative Inactivation and Bioremediation of Chloramphenicol.
Chembiochem, 24, 2023
|
|
3SHV
 
 | |
8BB4
 
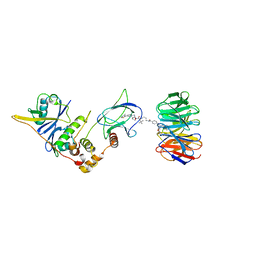 | | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with C3 linker | | Descriptor: | Elongin-B, Elongin-C, WD repeat-containing protein 5, ... | | Authors: | Kraemer, A, Doelle, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
1TV4
 
 | | Crystal structure of the sulfite MtmB complex | | Descriptor: | 3-METHYL-5-SULFO-PYRROLIDINE-2-CARBOXYLIC ACID, Monomethylamine methyltransferase mtmB1, SULFATE ION | | Authors: | Hao, B, Zhao, G, Kang, P.T, Soares, J.A, Ferguson, T.K, Gallucci, J, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2004-06-26 | | Release date: | 2004-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reactivity and chemical synthesis of L-pyrrolysine- the 22(nd) genetically encoded amino acid
Chem.Biol., 11, 2004
|
|
8BFD
 
 | | Racemic structure of PK-7 (310HD-U2U5) | | Descriptor: | 310HD-U2U5, D-310HD-U2U5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
1TV8
 
 | | Structure of MoaA in complex with S-adenosylmethionine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1TVE
 
 | | Homoserine Dehydrogenase in complex with 4-(4-hydroxy-3-isopropylphenylthio)-2-isopropylphenol | | Descriptor: | 4-(4-HYDROXY-3-ISOPROPYLPHENYLTHIO)-2-ISOPROPYLPHENOL, Homoserine dehydrogenase | | Authors: | Ejim, L, Mirza, I.A, Capone, C, Nazi, I, Jenkins, S, Chee, G.L, Berghuis, A.M, Wright, G.D. | | Deposit date: | 2004-06-29 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New phenolic inhibitors of yeast homoserine dehydrogenase
Bioorg.Med.Chem., 12, 2004
|
|
8B7J
 
 | | Human HSP90 alpha ATP Binding Domain, ATP-lid closed conformation, R46A | | Descriptor: | HSP90AA1 protein | | Authors: | Rioual, E, Henot, F, Favier, A, Macek, P, Crublet, E, Josso, P, Brustcher, B, Frech, M, Gans, P, Loison, C, Boisbouvier, J. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Visualizing the transiently populated closed-state of human HSP90 ATP binding domain.
Nat Commun, 13, 2022
|
|
8B7I
 
 | | Human HSP90 alpha ATP Binding Domain, ATP-lid open conformation, R60A | | Descriptor: | HSP90AA1 protein | | Authors: | Rioual, E, Henot, F, Favier, A, Macek, P, Crublet, E, Josso, P, Brutscher, B, Frech, M, Gans, P, Loison, C, Boisbouvier, J. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Visualizing the transiently populated closed-state of human HSP90 ATP binding domain.
Nat Commun, 13, 2022
|
|
1U3W
 
 | |
1U4G
 
 | | Elastase of Pseudomonas aeruginosa with an inhibitor | | Descriptor: | CALCIUM ION, Elastase, N-(1-CARBOXY-3-PHENYLPROPYL)PHENYLALANYL-ALPHA-ASPARAGINE, ... | | Authors: | Bitto, E, McKay, D.B. | | Deposit date: | 2004-07-25 | | Release date: | 2004-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Elastase of Pseudomonas aeruginosa with an inhibitor
To be Published, 2004
|
|
8AYK
 
 | | Crystal structure of D-amino acid aminotrensferase from Aminobacterium colombiense complexed with D-glutamate | | Descriptor: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
1U54
 
 | | Crystal Structures of the Phosphorylated and Unphosphorylated Kinase Domains of the CDC42-associated Tyrosine Kinase ACK1 bound to AMP-PCP | | Descriptor: | Activated CDC42 kinase 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Lougheed, J.C, Chen, R.H, Mak, P, Stout, T.J. | | Deposit date: | 2004-07-26 | | Release date: | 2004-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of the Phosphorylated and Unphosphorylated Kinase Domains of the Cdc42-associated Tyrosine Kinase ACK1.
J.Biol.Chem., 279, 2004
|
|
8BK0
 
 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with LDN-211904 | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chlorophenyl)-6-piperidin-4-yl-imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Zhubi, R, Gerninghaus, J, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with LDN-211904
To Be Published
|
|
1TW0
 
 | |
3SI2
 
 | | Structure of glycosylated murine glutaminyl cyclase in presence of the inhibitor PQ50 (PDBD150) | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Parthier, C, Carrillo, D, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
8BIN
 
 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with MR21 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-(2-chlorophenyl)-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Ephrin type-A receptor 2 | | Authors: | Zhubi, R, Rak, M, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Shifting the selectivity of pyrido[2,3-d]pyrimidin-7(8H)-one inhibitors towards the salt-inducible kinase (SIK) subfamily.
Eur.J.Med.Chem., 254, 2023
|
|
1TWZ
 
 | | Dihydropteroate Synthetase, With Bound Substrate Analogue PtP, From Bacillus anthracis | | Descriptor: | DHPS, Dihydropteroate synthase, PTERIN-6-YL-METHYL-MONOPHOSPHATE, ... | | Authors: | Babaoglu, K, Qi, J, Lee, R.L, White, S.W. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of 7,8-Dihydropteroate Synthase from Bacillus anthracis; Mechanism and Novel Inhibitor Design.
STRUCTURE, 12, 2004
|
|
3SIW
 
 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase co-crystallized with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
