7MIG
 
 | | Human CTPS1 bound to inhibitor T35 | | Descriptor: | 2-{2-[(cyclopropanesulfonyl)amino]-1,3-thiazol-4-yl}-2-methyl-N-{5-[6-(trifluoromethyl)pyrazin-2-yl]pyridin-2-yl}propanamide, CTP synthase 1, GLUTAMINE, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH0
 
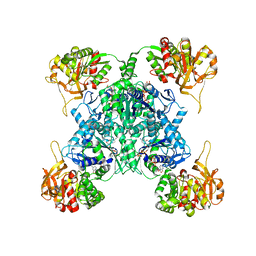 | | Human CTPS1 bound to CTP | | Descriptor: | CTP synthase 1, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Lynch, E.M, Kollman, J.M. | | Deposit date: | 2021-04-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIU
 
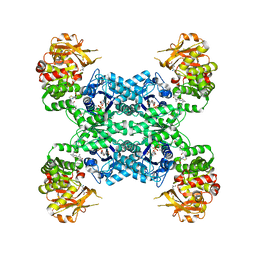 | | Mouse CTPS2 bound to inhibitor R80 | | Descriptor: | CTP synthase 2, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIH
 
 | | Human CTPS2 bound to inhibitor R80 | | Descriptor: | CTP synthase 2, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4FE2
 
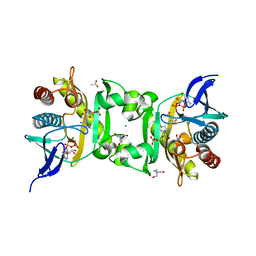 | | X-Ray Structure of SAICAR Synthetase (PurC) from Streptococcus pneumoniae complexed with AIR, ADP, Asp and Mg2+ | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wolf, N, Abad-Zapatero, C, Johnson, M.E, Fung, L.M.-W. | | Deposit date: | 2012-05-29 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structures of SAICAR synthetase (PurC) from Streptococcus pneumoniae with ADP, Mg(2+), AIR and Asp.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2VO1
 
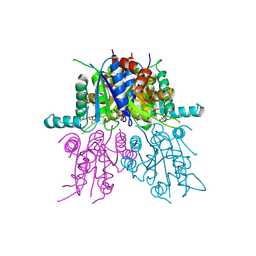 | | CRYSTAL STRUCTURE OF THE SYNTHETASE DOMAIN OF HUMAN CTP SYNTHETASE | | Descriptor: | CTP SYNTHASE 1, SULFATE ION | | Authors: | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Kotenyoa, T, Moche, M, Nilsson-Ehle, P, Ogg, D, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Nordlund, P. | | Deposit date: | 2008-02-08 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Synthetase Domain of Human Ctp Synthetase, a Target for Anticancer Therapy.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
4PL7
 
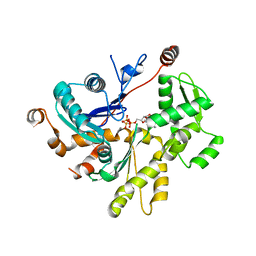 | |
4PL8
 
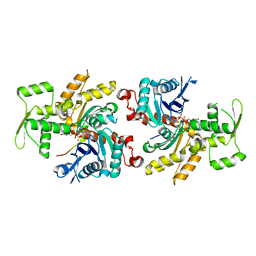 | |
4JLX
 
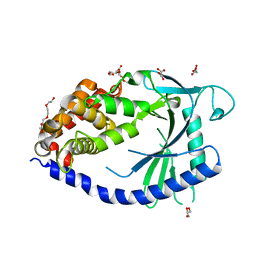 | |
4E4T
 
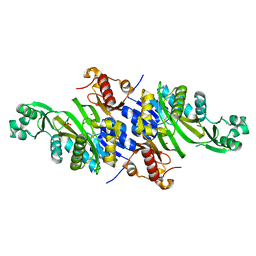 | |
6EEI
 
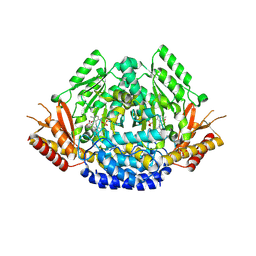 | | Crystal structure of Arabidopsis thaliana phenylacetaldehyde synthase in complex with L-phenylalanine | | Descriptor: | PHENYLALANINE, SULFATE ION, Tyrosine decarboxylase 1 | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-14 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (1.99001348 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7WXF
 
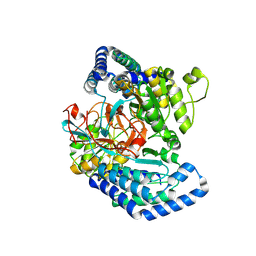 | |
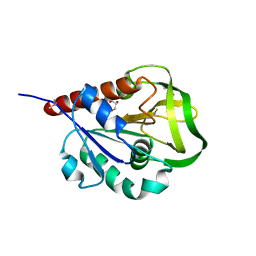
2A9V
 
 | |
4OIP
 
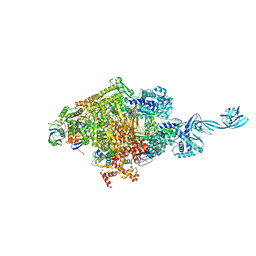 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
5D4C
 
 | |
2CG8
 
 | | The bifunctional dihydroneopterin aldolase 6-hydroxymethyl-7,8- dihydropterin synthase from Streptococcus pneumoniae | | Descriptor: | DIHYDRONEOPTERIN ALDOLASE 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN SYNTHASE | | Authors: | Garcon, A, Levy, C, Derrick, J.P. | | Deposit date: | 2006-02-28 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Bifunctional Dihydroneopterin Aldolase/6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase from Streptococcus Pneumoniae.
J.Mol.Biol., 360, 2006
|
|
4B2G
 
 | | Crystal Structure of an Indole-3-Acetic Acid Amido Synthase from Vitis vinifera Involved in Auxin Homeostasis | | Descriptor: | GH3-1 AUXIN CONJUGATING ENZYME, MALONATE ION, [(2S,3R,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl] 2-(1H-indol-3-yl)ethyl hydrogen phosphate | | Authors: | Peat, T.S, Bottcher, C, Newman, J, Lucent, D, Cowieson, N, Davies, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of an Indole-3-Acetic Acid Amido Synthetase from Grapevine Involved in Auxin Homeostasis.
Plant Cell, 24, 2012
|
|
3R9R
 
 | |
4OVM
 
 | | Crystal structure of SgcJ protein from Streptomyces carzinostaticus | | Descriptor: | uncharacterized protein SgcJ | | Authors: | Chang, C, Bigelow, L, Clancy, S, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.719 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J.Antibiot., 2016
|
|
4I4K
 
 | | Streptomyces globisporus C-1027 9-membered enediyne conserved protein SgcE6 | | Descriptor: | CITRIC ACID, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J Antibiot (Tokyo), 69, 2016
|
|
4NEO
 
 | | Structure of BlmI, a type-II acyl-carrier-protein from Streptomyces verticillus involved in bleomycin biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Peptide synthetase NRPS type II-PCP | | Authors: | Cuff, M.E, Bigelow, L, Bearden, J, Babnigg, G, Bruno, C.J.P, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of BlmI as a model for nonribosomal peptide synthetase peptidyl carrier proteins.
Proteins, 82, 2014
|
|
4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | Authors: | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
4HZP
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP Acyltransferase/Decarboxylase, CHLORIDE ION, COENZYME A, ... | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
4HZN
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional Methylmalonyl-CoA:ACP Acyltransferase/Decarboxylase, GLYCEROL, ... | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
4HZO
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP acyltransferase/decarboxylase, CHLORIDE ION, COENZYME A | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
