5NN8
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
1OSE
 
 | | Porcine pancreatic alpha-amylase complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Gilles, C, Payan, F. | | Deposit date: | 1996-03-20 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of pig pancreatic alpha-amylase isoenzyme II, in complex with the carbohydrate inhibitor acarbose.
Eur.J.Biochem., 238, 1996
|
|
5D4K
 
 | |
4BQF
 
 | | Arabidopsis thaliana cytosolic alpha-1,4-glucan phosphorylase (PHS2) in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA-GLUCAN PHOSPHORYLASE 2, CYTOSOLIC, ... | | Authors: | O'Neill, E.C, Rashid, A.M, Stevenson, C.E.M, Hetru, A.C, Gunning, A.P, Rejzek, M, Nepogodiev, S.A, Bornemann, S, Lawson, D.M, Field, R.A. | | Deposit date: | 2013-05-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Sugar-Coated Sensor Chip and Nanoparticle Surfaces for the in Vitro Enzymatic Synthesis of Starch-Like Materials
Chem.Sci., 5, 2014
|
|
6BOK
 
 | |
6B4V
 
 | |
6T15
 
 | | The III2-IV(5B)1 respiratory supercomplex from S. cerevisiae | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CARDIOLIPIN, COPPER (II) ION, ... | | Authors: | Marechal, A, Pinotsis, N, Hartley, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Rcf2 revealed in cryo-EM structures of hypoxic isoforms of mature mitochondrial III-IV supercomplexes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1HX0
 
 | | Structure of pig pancreatic alpha-amylase complexed with the "truncate" acarbose molecule (pseudotrisaccharide) | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ALPHA AMYLASE (PPA), ... | | Authors: | Qian, M, Payan, F. | | Deposit date: | 2001-01-11 | | Release date: | 2001-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Enzyme-catalyzed condensation reaction in a mammalian alpha-amylase. High-resolution structural analysis of an enzyme-inhibitor complex
Biochemistry, 40, 2001
|
|
6XKV
 
 | | R. capsulatus cyt bc1 with both FeS proteins in b position (CIII2 b-b) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XI0
 
 | | R. capsulatus cyt bc1 (CIII2) at 3.3A | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6XKT
 
 | | R. capsulatus cyt bc1 with both FeS proteins in c position (CIII2 c-c) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
4CPC
 
 | |
6XKU
 
 | | R. capsulatus cyt bc1 with one FeS protein in b position and one in c position (CIII2 b-c) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
1W9X
 
 | | Bacillus halmapalus alpha amylase | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA AMYLASE, CALCIUM ION, ... | | Authors: | Davies, G.J, Brzozowski, A.M, Dauter, Z, Rasmussen, M.D, Borchert, T.V, Wilson, K.S. | | Deposit date: | 2004-10-20 | | Release date: | 2005-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Bacillus Halmapalus Family 13 Alpha-Amylase, Bha, in Complex with an Acarbose-Derived Nonasaccharide at 2.1 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8PAY
 
 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 2. | | Descriptor: | ACE-GLN-ALC-GLC-LEU-PHE, Beta sliding clamp, GLYCEROL, ... | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, wagner, J, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
8PAT
 
 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 3. | | Descriptor: | ACE-GLN-ALC-GLX-LEU-PHE, Beta sliding clamp | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
8ORP
 
 | | Crystal structure of Drosophila melanogaster alpha-amylase in complex with the inhibitor acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Characterization of Drosophila melanogaster alpha-Amylase.
Molecules, 28, 2023
|
|
1C3O
 
 | | CRYSTAL STRUCTURE OF THE CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT MUTANT C269S WITH BOUND GLUTAMINE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT, CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-07-28 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Biochemistry, 38, 1999
|
|
5E8L
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase 11 from Arabidopsis thaliana | | Descriptor: | Heterodimeric geranylgeranyl pyrophosphate synthase large subunit 1, chloroplastic | | Authors: | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
7SB4
 
 | | Structure of OC43 spike in complex with polyclonal Fab2 (Donor 1412) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SBX
 
 | | Structure of OC43 spike in complex with polyclonal Fab6 (Donor 1051) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SBY
 
 | | Structure of OC43 spike in complex with polyclonal Fab7 (Donor 269) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SB5
 
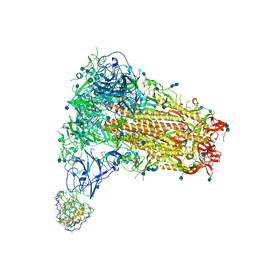 | | Structure of OC43 spike in complex with polyclonal Fab3 (Donor 1412) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SB3
 
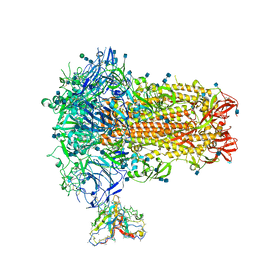 | | Structure of OC43 spike in complex with polyclonal Fab1 (Donor 269) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Ward, A, Bangaru, S, Antanasijevic, A. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SBW
 
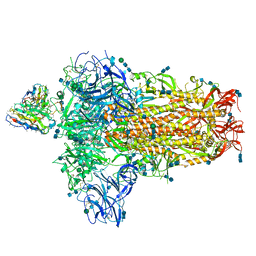 | | Structure of OC43 spike in complex with polyclonal Fab5 (Donor 1051) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
