1XED
 
 | |
1XT5
 
 | | Crystal Structure of VCBP3, domain 1, from Branchiostoma floridae | | Descriptor: | SULFATE ION, variable region-containing chitin-binding protein 3 | | Authors: | Hernandez Prada, J.A, Haire, R.N, Cannon, J.P, Allaire, M, Jakoncic, J, Stojanoff, V, Litman, G.W, Ostrov, D.A. | | Deposit date: | 2004-10-21 | | Release date: | 2005-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ancient evolutionary origin of diversified variable regions demonstrated by crystal structures of an immune-type receptor in amphioxus.
Nat.Immunol., 7, 2006
|
|
1URL
 
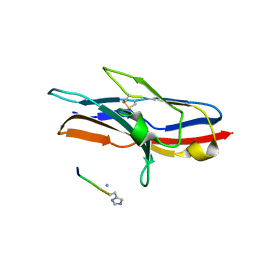 | | N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) IN COMPLEX WITH GLYCOPEPTIDE | | Descriptor: | ALA-GLY-HIS-THR-TRP-GLY-HIA, N-acetyl-alpha-neuraminic acid, SIALOADHESIN | | Authors: | Bukrinsky, J.T, Hilaire, P.M.S, Meldal, M, Crocker, P.R, Henriksen, A. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Complex of Sialoadhesin with a Glycopeptide Ligand
Biochim.Biophys.Acta, 1702, 2004
|
|
1VFA
 
 | |
3KHQ
 
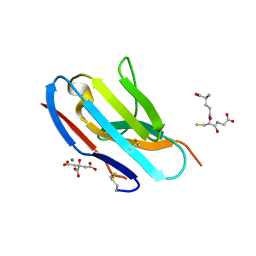 | | Crystal structure of murine Ig-beta (CD79b) in the monomeric form | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain, CITRATE ANION, GLUTATHIONE, ... | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-30 | | Release date: | 2010-08-25 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
5JXE
 
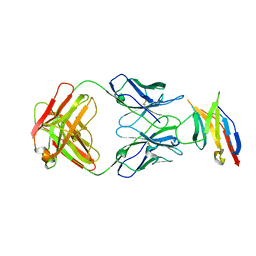 | | Human PD-1 ectodomain complexed with Pembrolizumab Fab | | Descriptor: | Pembrolizumab Fab heavy chain, Pembrolizumab Fab light chain, Programmed cell death protein 1 | | Authors: | Na, Z, Bharath, S.R, Song, H. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for blocking PD-1-mediated immune suppression by therapeutic antibody pembrolizumab.
Cell Res., 27, 2017
|
|
3KG5
 
 | | Crystal structure of human Ig-beta homodimer | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-28 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
6PLL
 
 | | Crystal structure of the ZIG-8 IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Zwei Ig domain protein zig-8 | | Authors: | Cheng, S, Ozkan, E. | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3KHO
 
 | | Crystal structure of murine Ig-beta (CD79b) homodimer | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain, SULFATE ION | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
3LVE
 
 | |
8EQ6
 
 | | PD1 signaling receptor bound to FAB Complex | | Descriptor: | Antibody FAB heavy chain, Antibody FAB light chain, Programmed cell death protein 1 | | Authors: | Bjorkelid, C, Paluch, C, Robertson, N.J. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Antibody agonists trigger immune receptor signaling through local exclusion of receptor-type protein tyrosine phosphatases.
Immunity, 57, 2024
|
|
6ON9
 
 | | Crystal Structure of the ZIG-8-RIG-5 IG1-IG1 heterodimer, tetragonal form | | Descriptor: | NeuRonal IgCAM-5, SODIUM ION, SULFATE ION, ... | | Authors: | Cheng, S, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5LVE
 
 | |
6ONB
 
 | | Crystal Structure of the ZIG-8-RIG-5 IG1-IG1 heterodimer, monoclinic form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NeuRonal IgCAM-5, ... | | Authors: | Cheng, S, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5MJ0
 
 | |
3MJ6
 
 | |
6ORT
 
 | | Crystal Structure of Bos taurus Mxra8 Ectodomain | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Matrix remodeling-associated protein 8 | | Authors: | Fremont, D.H, Kim, A.S, Nelson, C.A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Evolutionary Insertion in the Mxra8 Receptor-Binding Site Confers Resistance to Alphavirus Infection and Pathogenesis.
Cell Host Microbe, 27, 2020
|
|
3MFG
 
 | |
8EW6
 
 | | Anti-human CD8 VHH complex with CD8 alpha | | Descriptor: | Anti-CD8 alpha VHH, T-cell surface glycoprotein CD8 alpha chain | | Authors: | Kiefer, J.R, Williams, S, Davies, C.W, Koerber, J.T, Sriraman, S.K, Yin, Y.P. | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Development of an 18 F-labeled anti-human CD8 VHH for same-day immunoPET imaging.
Eur J Nucl Med Mol Imaging, 50, 2023
|
|
3MCG
 
 | |
3MJ9
 
 | |
5MJ1
 
 | | Extracellular domain of human CD83 - rhombohedral crystal form | | Descriptor: | CD83 antigen, DI(HYDROXYETHYL)ETHER | | Authors: | Klingl, S, Egerer-Sieber, C, Schmid, B, Weiler, S, Muller, Y.A. | | Deposit date: | 2016-11-29 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Extracellular Domain of the Human Dendritic Cell Surface Marker CD83.
J. Mol. Biol., 429, 2017
|
|
5MJ2
 
 | | Extracellular domain of human CD83 - rhombohedral crystal form after UV-RIP (S-SAD data) | | Descriptor: | CD83 antigen, DI(HYDROXYETHYL)ETHER | | Authors: | Klingl, S, Egerer-Sieber, C, Schmid, B, Weiler, S, Muller, Y.A. | | Deposit date: | 2016-11-29 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of the Extracellular Domain of the Human Dendritic Cell Surface Marker CD83.
J. Mol. Biol., 429, 2017
|
|
3M45
 
 | | Crystal structure of Ig1 domain of mouse SynCAM 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 2 | | Authors: | Yue, L, Modis, Y. | | Deposit date: | 2010-03-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | N-glycosylation at the SynCAM (synaptic cell adhesion molecule) immunoglobulin interface modulates synaptic adhesion.
J.Biol.Chem., 285, 2010
|
|
3NOI
 
 | | Crystal Structure of Natural Killer Cell Cytotoxicity Receptor NKp30 (NCR3) | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CALCIUM ION, Natural cytotoxicity triggering receptor 3 | | Authors: | Joyce, M.G, Sun, P.D. | | Deposit date: | 2010-06-25 | | Release date: | 2011-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Structure of the Natural Killer cell activating receptor NKp30 and dissection of ligand binding site
Proc.Natl.Acad.Sci.USA, 2011
|
|
