5VAI
 
 | | Cryo-EM structure of the activated Glucagon-like peptide-1 receptor in complex with G protein | | Descriptor: | Glucagon-like peptide 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, Y, Sun, B, Feng, D, Hu, H, Chu, M, Qu, Q, Tarrasch, J.T, Li, S, Kobilka, T.S, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2017-03-27 | | Release date: | 2017-05-24 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein.
Nature, 546, 2017
|
|
4MU0
 
 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+ and 1,2,4-triazole at 1.3 A resolution | | Descriptor: | 1,2,4-TRIAZOLE, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
3ZW9
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH (2S,3S)-3-HYDROXY-2-METHYLBUTANOYL-COA | | Descriptor: | (2S,3S)-3-HYDROXY-2-METHYLBUTANOYL-COA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PEROXISOMAL BIFUNCTIONAL ENZYME, ... | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
2F3A
 
 | |
7KVK
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450 3A4, GLYCEROL, ... | | Authors: | Sevrioukova, I. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Rational Design of CYP3A4 Inhibitors: A One-Atom Linker Elongation in Ritonavir-Like Compounds Leads to a Marked Improvement in the Binding Strength.
Int J Mol Sci, 22, 2021
|
|
5OTT
 
 | |
4CN9
 
 | | structure of proximal thread matrix protein 1 (PTMP1) from the mussel byssus with zinc occupied MIDAS motif | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PROXIMAL THREAD MATRIX PROTEIN 1, ... | | Authors: | Gertz, M, Suhre, M.H, Scheibel, T, Steegborn, C. | | Deposit date: | 2014-01-21 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Features of a Collagen-Binding Matrix Protein from the Mussel Byssus.
Nat.Commun., 5, 2014
|
|
4KK2
 
 | | Crystal structure of a chimeric FPP/GFPP synthase (TARGET EFI-502313c) from Artemisia spiciformiS (1-72:GI751454468,73-346:GI75233326), apo structure | | Descriptor: | Monoterpene synthase FDS-5, chloroplastic - Farnesyl diphosphate synthase 1 chimera | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-05-05 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a chimeric FPP/GFPP synthase (TARGET EFI-502313c) from Artemisia spiciformis (1-72:GI751454468,73-346:GI75233326), apo structure
To be Published
|
|
1WDD
 
 | | Crystal Structure of Activated Rice Rubisco Complexed with 2-Carboxyarabinitol-1,5-bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mizohata, E, Matsumura, H, Ueno, T, Ishida, H, Inoue, T, Makino, A, Mae, T, Kai, Y. | | Deposit date: | 2004-05-13 | | Release date: | 2004-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
1WMF
 
 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (oxidized form, 1.73 angstrom) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
4CTM
 
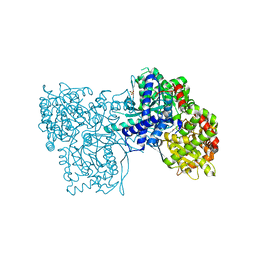 | | Glucopyranosylidene-spiro-iminothiazolidinone, a New Bicyclic Ring System: Synthesis, Derivatization, and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetic and Crystallographic Methods | | Descriptor: | (5R,7R,8S,9S,10R)-8,9,10-trihydroxy-7-(hydroxymethyl)-2-imino-6-oxa-1-thia-3-azaspiro[4.5]decan-4-one, GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, ... | | Authors: | Alexacou, K.M, Papakonstantinou, M, Leonidas, D.D, Zographos, S.E, Chrysina, E.D. | | Deposit date: | 2014-03-15 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Glucopyranosylidene-Spiro-Iminothiazolidinone, a New Bicyclic Ring System: Synthesis, Derivatization, and Evaluation for Inhibition of Glycogen Phosphorylase by Enzyme Kinetic and Crystallographic Methods.
Bioorg.Med.Chem., 22, 2014
|
|
3UV4
 
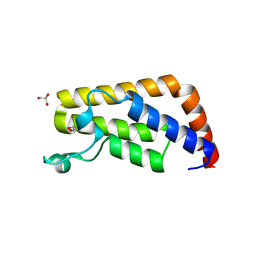 | | Crystal Structure of the second bromodomain of human Transcription initiation factor TFIID subunit 1 (TAF1) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3UVJ
 
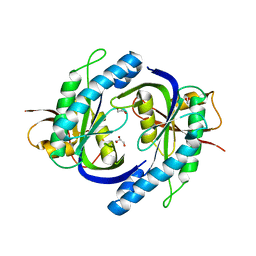 | | Crystal structure of the catalytic domain of the heterodimeric human soluble guanylate cyclase 1. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Guanylate cyclase soluble subunit alpha-3, ... | | Authors: | Allerston, C.K, Berridge, G, Chalk, R, Cooper, C.D.O, Savitsky, P, Vollmar, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of the catalytic domain of human soluble guanylate cyclase.
Plos One, 8, 2013
|
|
2FP2
 
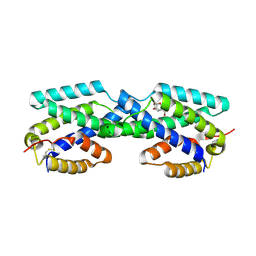 | | Secreted Chorismate Mutase from Mycobacterium tuberculosis | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, Chorismate mutase | | Authors: | Okvist, M, Dey, R, Sasso, S, Grahn, E, Kast, P, Krengel, U. | | Deposit date: | 2006-01-15 | | Release date: | 2006-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | 1.6A Crystal Structure of the Secreted Chorismate Mutase from Mycobacterium tuberculosis: Novel Fold Topology Revealed
J.Mol.Biol., 357, 2006
|
|
1RVG
 
 | |
2QJI
 
 | | M. jannaschii ADH synthase complexed with dihydroxyacetone phosphate and glycerol | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, GLYCEROL, Putative aldolase MJ0400 | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
Biochemistry, 46, 2007
|
|
1G8K
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ARSENITE OXIDASE FROM ALCALIGENES FAECALIS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ARSENITE OXIDASE, ... | | Authors: | Ellis, P.J, Conrads, T, Hille, R, Kuhn, P. | | Deposit date: | 2000-11-17 | | Release date: | 2000-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A.
Structure, 9, 2001
|
|
4D6F
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A, X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6I
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4Q0O
 
 | | Crystal Structure of the fifth bromodomain of Human Poly-bromodomain containing protein 1 (PB1) in complex with a hydroxyphenyl-propenone ligand | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-[(3R)-3-phenylpiperidin-1-yl]prop-2-en-1-one, POTASSIUM ION, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of the fifth bromodomain of Human Poly-bromodomain containing protein 1 (PB1) in complex with a hydroxyphenyl-propenone ligand
To be Published
|
|
4D6H
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
5YVN
 
 | | Human Glutathione Transferase Omega1 | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase omega-1, ... | | Authors: | Saisawang, C, Ketterman, A, Wongsantichon, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Glutathione transferase Omega 1-1 (GSTO1-1) modulates Akt and MEK1/2 signaling in human neuroblastoma cell SH-SY5Y.
Proteins, 87, 2019
|
|
5PZK
 
 | |
1RPI
 
 | | Crystal structures of a Multidrug-Resistant HIV-1 Protease Reveal an Expanded Active Site Cavity | | Descriptor: | alpha-D-glucopyranose, protease | | Authors: | Logsdon, B.C, Vickrey, J.F, Martin, P, Proteasa, G, Koepke, J.I, Terlecky, S.R, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2003-12-03 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity.
J.Virol., 78, 2004
|
|
4HJ2
 
 | | Crystal Structure Analysis of GSTA1-1 in complex with chlorambucil | | Descriptor: | Glutathione S-transferase A1, L-gamma-glutamyl-S-(2-{[4-(3-carboxypropyl)phenyl](2-chloroethyl)amino}ethyl)-L-cysteinylglycine | | Authors: | Karpusas, M, Chiniadis, L, Labrou, N.E. | | Deposit date: | 2012-10-12 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The interaction of the chemotherapeutic drug chlorambucil with human glutathione transferase A1-1: kinetic and structural analysis.
Plos One, 8, 2013
|
|
