4QR9
 
 | | Crystal structure of two HMGB1 Box A domains cooperating to underwind and kink a DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*GP*AP*TP*AP*T)-3'), High mobility group protein B1, MAGNESIUM ION | | Authors: | Sanchez-Giraldo, R, Acosta-Reyes, F.J, Malarkey, C.S, Saperas, N, Churchill, M.E.A, Campos, J.L. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two high-mobility group box domains act together to underwind and kink DNA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1LV5
 
 | | Crystal Structure of the Closed Conformation of Bacillus DNA Polymerase I Fragment Bound to DNA and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*CP*GP*TP*CP*GP*CP*TP*GP*AP*TP*CP*CP*G)-3', 5'-D(*GP*GP*AP*TP*CP*AP*GP*CP*GP*A)-3', ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-05-24 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3U58
 
 | | Crystal Structure of the Tetrahymena telomerase processivity factor Teb1 AB | | Descriptor: | DNA (5'-D(*GP*GP*GP*T)-3'), Tetrahymena Teb1 AB | | Authors: | Zeng, Z, Huang, J, Yang, Y, Lei, M. | | Deposit date: | 2011-10-11 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1YSA
 
 | | THE GCN4 BASIC REGION LEUCINE ZIPPER BINDS DNA AS A DIMER OF UNINTERRUPTED ALPHA HELICES: CRYSTAL STRUCTURE OF THE PROTEIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*CP*TP*GP*GP*AP*TP*GP*AP*GP*TP*CP*AP*TP*A P*GP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*AP*TP*CP*CP*A P*GP*TP*T)-3'), PROTEIN (GCN4) | | Authors: | Ellenberger, T.E, Brandl, C.J, Struhl, K, Harrison, S.C. | | Deposit date: | 1993-08-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex.
Cell(Cambridge,Mass.), 71, 1992
|
|
2I9G
 
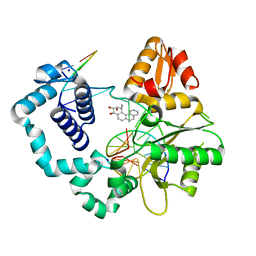 | | DNA Polymerase Beta with a Benzo[c]phenanthrene diol epoxide adducted guanine base | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Batra, V.K, Wilson, S.H, Beard, W.A, Pedersen, L.C. | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of DNA polymerase beta with a benzo[c]phenanthrene diol epoxide-adducted template exhibits mutagenic features.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DGC
 
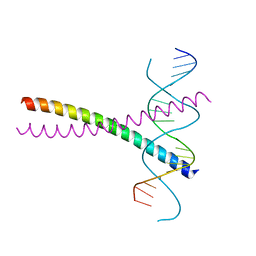 | |
6R9B
 
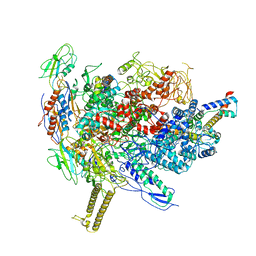 | |
4XRH
 
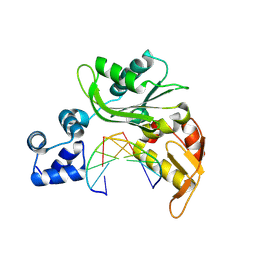 | |
2JGU
 
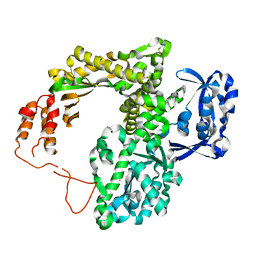 | | crystal structure of DNA-directed DNA polymerase | | Descriptor: | DNA POLYMERASE, MANGANESE (II) ION | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2007-02-15 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Pfu, the High Fidelity DNA Polymerase from Pyrococcus Furiosus.
Int.J.Biol.Macromol., 42, 2008
|
|
7DCJ
 
 | |
7DCT
 
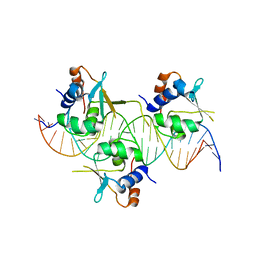 | | Crystal structure of HSF1 DNA-binding domain in complex with 3-site HSE DNA (24 bp) | | Descriptor: | DNA (5'-D(*AP*CP*TP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*AP*G)-3'), Heat shock factor protein 1, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
7DCU
 
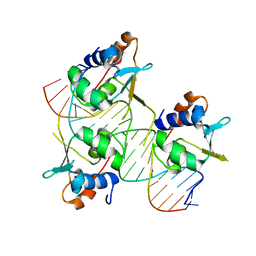 | | Crystal structure of HSF2 DNA-binding domain in complex with 3-site HSE DNA (21 bp) | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*G)-3'), Heat shock factor protein 2, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
7DCS
 
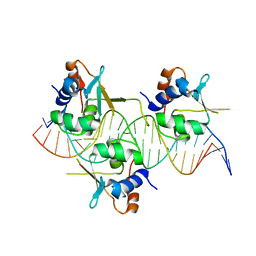 | | Crystal structure of HSF1 DNA-binding domain in complex with 3-site HSE DNA (23 bp) | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*GP*A)-3'), Heat shock factor protein 1, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
8Q3R
 
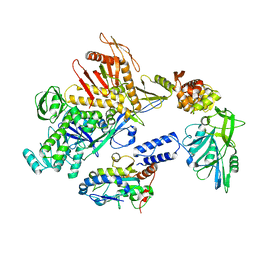 | | Cryo-EM structure of the DNA polymerase holoenzyme E9-A20-D4 of vaccinia virus | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component OPG148, Uracil-DNA glycosylase | | Authors: | Burmeister, W.P, Ballandras-Colas, A, Boettcher, B, Grimm, C. | | Deposit date: | 2023-08-04 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and flexibility of the DNA polymerase holoenzyme of vaccinia virus.
Plos Pathog., 20, 2024
|
|
3MKY
 
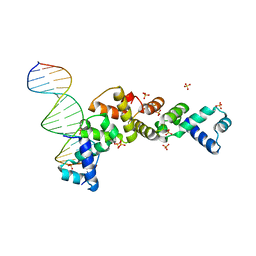 | | Structure of SopB(155-323)-18mer DNA complex, I23 form | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | Authors: | Schumacher, M.A, Piro, K, Xu, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
6R9G
 
 | |
1L3T
 
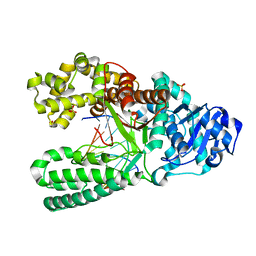 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 10 base pairs of duplex DNA following addition of a single dTTP residue | | Descriptor: | 5'-D(*GP*AP*CP*G*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8EFC
 
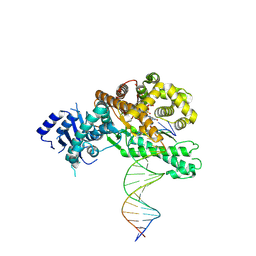 | | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*TP*GP*TP*GP*AP*GP*GP*CP*AP*TP*CP*CP*GP*TP*AP*GP*(2DA))-3'), DNA (5'-D(*AP*GP*CP*TP*CP*TP*AP*CP*GP*GP*AP*TP*GP*CP*CP*TP*CP*AP*CP*AP*G)-3'), ... | | Authors: | Li, C, Zhu, H, Sun, J, Gao, Y. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of DNA polymerase theta mediated DNA end joining.
Nucleic Acids Res., 51, 2023
|
|
1SKN
 
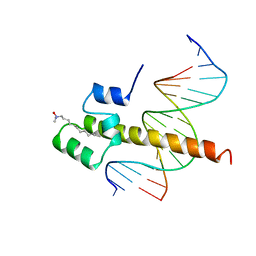 | | THE BINDING DOMAIN OF SKN-1 IN COMPLEX WITH DNA: A NEW DNA-BINDING MOTIF | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*GP*AP*TP*GP*AP*CP*AP*TP*TP*GP*T)-3'), DNA (5'-D(*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*TP*CP*CP*C)-3'), DNA-BINDING DOMAIN OF SKN-1, ... | | Authors: | Rupert, P.B, Daughdrill, G.W, Bowerman, B, Matthews, B.W. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new DNA-binding motif in the Skn-1 binding domain-DNA complex.
Nat.Struct.Biol., 5, 1998
|
|
2JXI
 
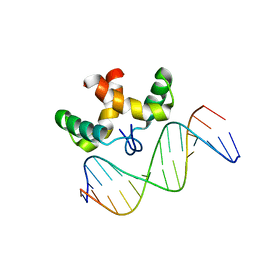 | | Solution structure of the DNA-binding domain of Pseudomonas putida Proline utilization A (putA) bound to GTTGCA DNA sequence | | Descriptor: | DNA (5'-D(*DAP*DAP*DAP*DGP*DGP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DC)-3'), DNA (5'-D(*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DCP*DTP*DTP*DT)-3'), Proline dehydrogenase | | Authors: | Halouska, S, Zhou, Y, Becker, D, Powers, R. | | Deposit date: | 2007-11-19 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Pseudomonas putida protein PpPutA45 and its DNA complex
Proteins, 75, 2008
|
|
6T8H
 
 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | Descriptor: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
2ATQ
 
 | | RB69 single-stranded DNA binding protein-DNA polymerase fusion | | Descriptor: | DNA polymerase, GUANOSINE-5'-DIPHOSPHATE, ZINC ION, ... | | Authors: | Sun, S, Geng, L, Shamoo, Y. | | Deposit date: | 2005-08-25 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and enzymatic properties of a chimeric bacteriophage RB69 DNA polymerase and single-stranded DNA binding protein with increased processivity.
Proteins, 65, 2006
|
|
6XTX
 
 | | CryoEM structure of human CMG bound to ATPgammaS and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45 homolog, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Rzechorzek, N.J, Pellegrini, L. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-27 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | CryoEM structures of human CMG-ATP gamma S-DNA and CMG-AND-1 complexes.
Nucleic Acids Res., 48, 2020
|
|
5FKU
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.34 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
7E92
 
 | |
