6PFX
 
 | |
6FNH
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with a pyrazolo[3,4-d]pyrimidine fragment of NVP-BHG712 | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-amine, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
6UPE
 
 | |
6FNK
 
 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase with a pyrazolo[3,4-d]pyrimidine fragment of NVP-BHG712 | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-amine, Ephrin type-B receptor 4 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.049 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
8DG6
 
 | |
7PMT
 
 | | Human Cyclophilin D in complex with N-[(5-ethyl-4-oxo-1,2,3,4,5,6- hexahydro-1,5-benzodiazocin-8-yl)methyl]-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide | | Descriptor: | DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ... | | Authors: | Silva, D.O, Graedler, U, Bandeiras, T.M. | | Deposit date: | 2021-09-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Human Cyclophilin D in complex with N-[(4-aminophenyl)methyl]-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide
To Be Published
|
|
6X9B
 
 | |
8PXM
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH (1R,1'R)-7,7'-(pentane-1,5-diylbis(oxy))bis(1,3-dimethyl-1,3-dihydro-2H-benzo[d]azepin-2-one) | | Descriptor: | (1R)-7-[5-[[(1R)-1,3-dimethyl-2-oxidanylidene-1H-3-benzazepin-7-yl]oxy]pentoxy]-1,3-dimethyl-1H-3-benzazepin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Chung, C.W. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.378 Å) | | Cite: | Design and Characterization of 1,3-Dihydro-2 H -benzo[ d ]azepin-2-ones as Rule-of-5 Compliant Bivalent BET Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
4X9U
 
 | | Crystal structure of the kiwifruit allergen Act d 5 | | Descriptor: | Kiwellin | | Authors: | Offermann, L.R, Perdue, M.L, Giangrieco, I, Tamburrini, M, Ciardiello, M.A, Chruszcz, M. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elusive Structural, Functional, and Immunological Features of Act d 5, the Green Kiwifruit Kiwellin.
J.Agric.Food Chem., 63, 2015
|
|
7A1E
 
 | | LppS with covalent adduct derived from 1c | | Descriptor: | ACETATE ION, L,D-transpeptidase 2, phenylmethanethiol | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-12 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
7A10
 
 | | LppS with covalent adduct derived from 1g | | Descriptor: | 4-methoxycyclohexa-2,5-diene-1-thione, L,D-transpeptidase 2 | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
7A11
 
 | | LppS with covalent adduct derived from 1E | | Descriptor: | ACETATE ION, L,D-transpeptidase 2, propane-1-thiol | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
7A0Z
 
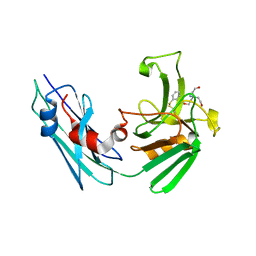 | | LppS with covalent adduct derived from 1b | | Descriptor: | L,D-transpeptidase 2, TRIS(HYDROXYETHYL)AMINOMETHANE, benzenethiol | | Authors: | Schnell, R, Steiner, E.M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | N-Thio-beta-lactams targeting L,D-transpeptidase-2, with activity against drug-resistant strains of Mycobacterium tuberculosis.
Cell Chem Biol, 28, 2021
|
|
7A1C
 
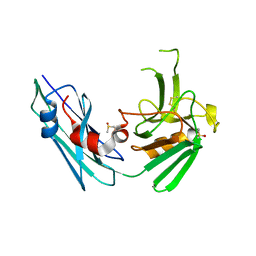 | |
7AEN
 
 | | Galectin-8 N-terminal carbohydrate recognition domain in complex with methyl 3-O-((7-carboxy)quinolin-2-yl)-methoxy)-beta-D-galactopyranoside | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoform 2 of Galectin-8, ... | | Authors: | Hassan, M, Klavern, V.S, Hakansson, M, Anderluh, M, Tomasic, T, Jakopin, Z, Nilsson, J.U, Kovacic, R, Walse, B, Diehl, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of d-Galactal Derivatives with High Affinity and Selectivity for the Galectin-8 N-Terminal Domain
Acs Med.Chem.Lett., 12, 2021
|
|
7Q3S
 
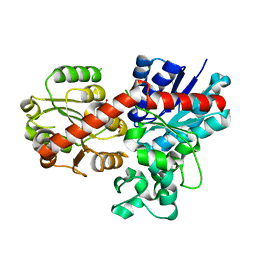 | | Crystal structure of UGT706F8 from Zea mays | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Fredslund, F, Bidart, G.N, Welner, D.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Family 1 Glycosyltransferase UGT706F8 from Zea mays Selectively Catalyzes the Synthesis of Silibinin 7- O -beta-d-Glucoside.
Acs Sustain Chem Eng, 10, 2022
|
|
6NNC
 
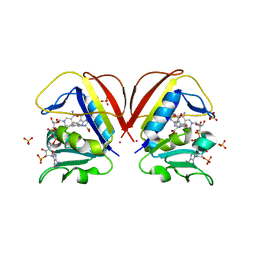 | | Structure of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with NADPH and pemetrexed | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8H4E
 
 | | Blasnase-T13A/P55N with D-asn | | Descriptor: | D-ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
1B3P
 
 | | 5'-D(*GP*GP*AP*GP*GP*AP*T)-3' | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*AP*T)-3') | | Authors: | Kettani, A, Bouaziz, S, Skripkin, E, Majumdar, A, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1998-12-14 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Interlocked mismatch-aligned arrowhead DNA motifs.
Structure Fold.Des., 7, 1999
|
|
8H4F
 
 | | Blasnase-T13A/P55F with D-asn | | Descriptor: | D-ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
6Y6F
 
 | | Crystal structure of STK17B (DRAK2) in complex with PKIS43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-methylsulfanylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
6Y6H
 
 | | Crystal structure of STK17b (DRAK2) in complex with UNC-AP-194 probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(1-benzothiophen-2-yl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
6X9A
 
 | |
6NZ2
 
 | |
3FVE
 
 | | Crystal structure of diaminopimelate epimerase Mycobacterium tuberculosis DapF | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Diaminopimelate epimerase, GLYCEROL | | Authors: | Usha, V, Dover, L.G, Roper, D.I, Futterer, K, Besra, G.S. | | Deposit date: | 2009-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the diaminopimelate epimerase DapF from Mycobacterium tuberculosis
Acta Crystallogr.,Sect.D, 65, 2009
|
|
