7QCL
 
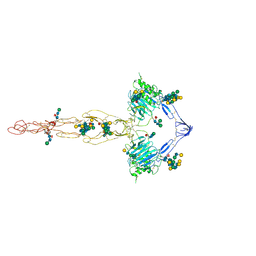 | | Structure of the MUCIN-2 Cterminal domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mucin-2, ... | | Authors: | Gallego, P, Hansson, G.C. | | Deposit date: | 2021-11-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The intestinal MUC2 mucin C-terminus is stabilized by an extra disulfide bond in comparison to von Willebrand factor and other gel-forming mucins.
Nat Commun, 14, 2023
|
|
3E6U
 
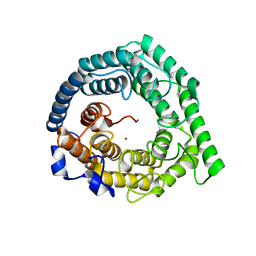 | | Crystal structure of Human LanCL1 | | Descriptor: | LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-16 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
6OSJ
 
 | |
8XF1
 
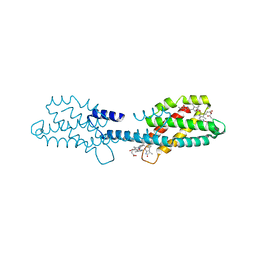 | | Crystal structure of the dissociated C-phycocyanin beta-chain from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Nguyen, H.K, Teramoto, T, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Disassembly and reassembly of the non-conventional thermophilic C-phycocyanin.
J.Biosci.Bioeng., 137, 2024
|
|
6XG1
 
 | | Class C beta-lactamase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Class C beta-lactamase from Escherichia coli
To Be Published
|
|
6YJC
 
 | | Crystal structure of p38alpha in complex with SR154 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-oxidanylidene-1-piperazin-1-yl-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-02 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74100935 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6ZWR
 
 | | p38a bound with SR92 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-oxidanylidene-1-(pyridin-4-ylmethylamino)butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Schroeder, M, Roehm, S, Knapp, S, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-28 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
7PPI
 
 | | Crystal STRUCTURE OF NAMPT IN COMPLEX WITH Compound 11 | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[4-[(5R)-1-(4-azanylbutyl)-6-oxidanylidene-5-quinolin-5-yl-4,5-dihydropyridazin-3-yl]phenyl]-1,3-dihydropyrrolo[3,4-c]pyridine-2-carboxamide, ... | | Authors: | Hillig, R.C. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A Novel NAMPT Inhibitor-Based Antibody-Drug Conjugate Payload Class for Cancer Therapy.
Bioconjug.Chem., 33, 2022
|
|
7ZS0
 
 | |
7ZS1
 
 | |
7ZS2
 
 | |
8W72
 
 | |
6YK7
 
 | | Crystal structure of p38 in complex with SR43 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(ethylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-05 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
8WCK
 
 | | FCP tetramer in Chaetoceros gracilis | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, CHLOROPHYLL A, Chlorophyll a/b-binding protein, ... | | Authors: | Feng, Y, Li, Z, Zhou, C, Shen, J.-R, Liu, C, Wang, W. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 5, 2024
|
|
6NZC
 
 | |
7PPG
 
 | | CRYSTAL STRUCTURE OF NAMPT IN COMPLEX WITH COMPOUND 9 | | Descriptor: | 1,2-ETHANEDIOL, N-[4-[(4R)-1-cyclopentyl-4-methyl-6-oxidanylidene-4,5-dihydropyridazin-3-yl]phenyl]-1,3-dihydropyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Hillig, R.C. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A Novel NAMPT Inhibitor-Based Antibody-Drug Conjugate Payload Class for Cancer Therapy.
Bioconjug.Chem., 33, 2022
|
|
6XVM
 
 | |
6ZWP
 
 | | p38a bound with SR348 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[(4-fluorophenyl)amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Schroeder, M, Roehm, S, Knapp, S, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-28 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6NZA
 
 | |
5COK
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-0476 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
8P2L
 
 | |
6Y3V
 
 | | 14-3-3 Sigma in complex with phosphorylated c-Jun peptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Ballone, A, Lau, R.A, Zweipfenning, F.P.A, Ottmann, C. | | Deposit date: | 2020-02-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | A new soaking procedure for X-ray crystallographic structural determination of protein-peptide complexes.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
9BCX
 
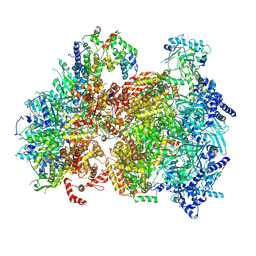 | | Cryo-EM structure of the S. cerevisiae ORC-Cdc6-Mcm2-7-DNA complex with a fully closed Mcm2-Mcm5 DNA entry gate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA (39-MER), ... | | Authors: | Yuan, Z, Bai, L, Li, H, Speck, C. | | Deposit date: | 2024-04-10 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | MCM2-7 ring closure involves the Mcm5 C-terminus and triggers Mcm4 ATP hydrolysis.
Nat Commun, 16, 2025
|
|
6NZB
 
 | |
5COP
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-097 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-(4-methoxyphenyl)butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
