8KC1
 
 | | De novo design protein -NX5 | | Descriptor: | De novo design protein -NX5 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design protein -NX5
To Be Published
|
|
8KC8
 
 | | De novo design protein -T11 | | Descriptor: | De novo design protein -T11 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo design protein -T11
To Be Published
|
|
8SBC
 
 | | Co-structure of Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform and brain penetrant inhibitors | | Descriptor: | (2M)-7-[(3R)-3-methylmorpholin-4-yl]-5-[(3S)-3-methylmorpholin-4-yl]-2-(pyridin-2-yl)-1H-imidazo[4,5-b]pyridine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Knapp, M.S, Elling, R.A, Tang, J. | | Deposit date: | 2023-04-03 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Brain-Penetrant ATP-Competitive mTOR Inhibitors for CNS Syndromes.
J.Med.Chem., 66, 2023
|
|
8KCV
 
 | | Crystal structure of UDA01-CAAMDDFQL | | Descriptor: | Beta2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of duck MHC UDA01
To Be Published
|
|
8KFO
 
 | | Crystal structure of BSA in complex with B3 | | Descriptor: | 6-[(~{E})-2-[1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyridin-1-ium-4-yl]ethenyl]-~{N},~{N}-dimethyl-naphthalen-2-amine, Albumin | | Authors: | Chen, X, Ge, Y.H, Yang, H, Fang, B, Li, L. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Bioinspired two-stage assembled photosensitive protein engineering for tumor-specific mitochondrial targeted phototherapy
To Be Published
|
|
5UDX
 
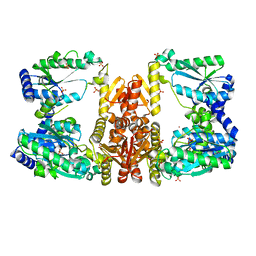 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with zinc | | Descriptor: | Lactate racemization operon protein LarE, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7P5R
 
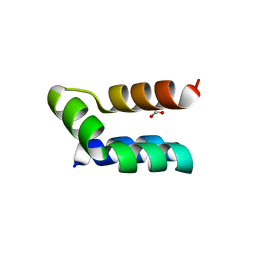 | | Racemic protein crystal structure of lacticin Q from Lactococcus lactis | | Descriptor: | D-Lacticin Q, FORMIC ACID, Lacticin Q | | Authors: | Lander, A.J, Li, X, Baumann, P, Rizkallah, P, Luk, L.Y.P, Jin, Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
8SB6
 
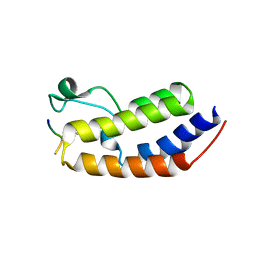 | | Structure of human BRD2-BD1 bound to a histone H4 acetyl-methyllysine peptide | | Descriptor: | Bromodomain containing 2, Histone H4 | | Authors: | Connor, L.J, Ekundayo, B.E, Lu-Culligan, W.J, Simon, M.D, Bleichert, F. | | Deposit date: | 2023-04-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acetyl-methyllysine marks chromatin at active transcription start sites.
Nature, 622, 2023
|
|
7YH4
 
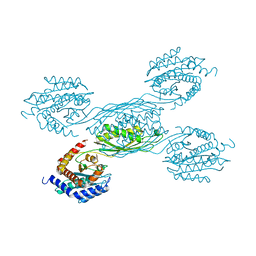 | |
8KA7
 
 | | De novo design protein -NB7 | | Descriptor: | De novo design protein -NB7 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo design protein -NB7
To Be Published
|
|
8S9N
 
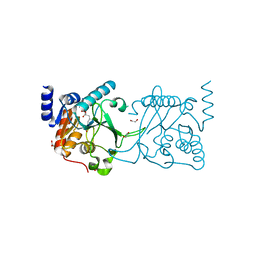 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - C2 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
6MVH
 
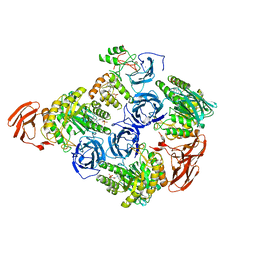 | |
8KBF
 
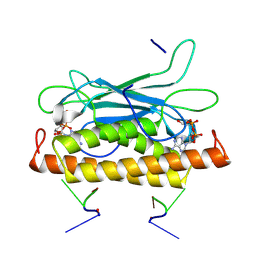 | | Structure of CbTad1 complexed with 1',3'-cADPR and cA3 | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Thoeris anti-defense 1 | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of CbTad1 complexed with 1',3'-cADPR and cA3
To Be Published
|
|
8S9O
 
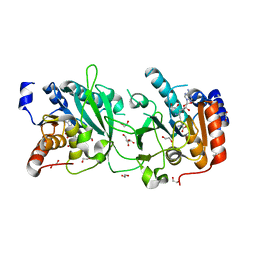 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - P1 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
6MJ8
 
 | |
8KDL
 
 | |
5GW8
 
 | | Crystal structure of a putative DAG-like lipase (MgMDL2) from Malassezia globosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Xu, J, Xu, H, Liu, J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Malassezia globosa MgMDL2 lipase: Crystal structure and rational modification of substrate specificity.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
7VSW
 
 | |
8UKZ
 
 | |
7P2U
 
 | | Crystal structure of Schistosoma mansoni HDAC8 in complex with a 3-chlorophenyl-spiroindoline capped hydroxamate-based inhibitor, bound to a novel site | | Descriptor: | 5-[[(2R)-2-(3-chlorophenyl)-1'-methyl-spiro[2H-indole-3,4'-piperidine]-1-yl]methyl]-N-oxidanyl-thiophene-2-carboxamide, CHLORIDE ION, Histone deacetylase 8, ... | | Authors: | Saccoccia, F, Gemma, S, Campiani, G, Ruberti, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Schistosoma mansoni histone deacetylase 8 reveal a novel binding site for allosteric inhibitors.
J.Biol.Chem., 298, 2022
|
|
7P3L
 
 | |
5C3T
 
 | | PD-1 binding domain from human PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Dubin, G, Holak, T.A. | | Deposit date: | 2015-06-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1.
Structure, 23, 2015
|
|
8KBK
 
 | | Structure of AcrIIA7 complexed with 1',2'-cADPR and cGG | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of AcrIIA7 complexed with 1',2'-cADPR and cGG
To Be Published
|
|
5TVW
 
 | | Crystal structure of mitochondrial Hsp90 (TRAP1) with ATP in absence of Mg, hemi-hydrolyzed | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Elnatan, D, Betegon, M, Agard, D.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-11-10 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Symmetry broken and rebroken during the ATP hydrolysis cycle of the mitochondrial Hsp90 TRAP1.
Elife, 6, 2017
|
|
6MKZ
 
 | | Crystal structure of murine 4-1BB/4-1BBL complex | | Descriptor: | Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bitra, A, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-09-26 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the m4-1BB/4-1BBL complex reveals an unusual dimeric ligand that undergoes structural changes upon 4-1BB receptor binding.
J. Biol. Chem., 294, 2019
|
|
