8H03
 
 | |
8H04
 
 | |
7OLA
 
 | | Structure of Primase-Helicase in SaPI5 | | Descriptor: | DNA primase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Qiao, C.C, Mir-Sanchis, I. | | Deposit date: | 2021-05-19 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Staphylococcal self-loading helicases couple the staircase mechanism with inter domain high flexibility.
Nucleic Acids Res., 50, 2022
|
|
4UZC
 
 | |
8HKW
 
 | |
4UX8
 
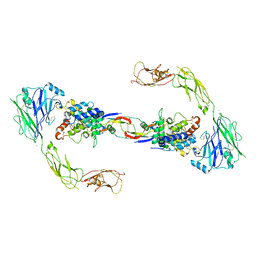 | | RET recognition of GDNF-GFRalpha1 ligand by a composite binding site promotes membrane-proximal self-association | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF FAMILY RECEPTOR ALPHA-1, ... | | Authors: | Goodman, K, Kjaer, S, Beuron, F, Knowles, P, Nawrotek, A, Burns, E, Purkiss, A, George, R, Santoro, M, Morris, E.P, McDonald, N.Q. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Ret Recognition of Gdnf-Gfralpha1 Ligand by a Composite Binding Site Promotes Membrane-Proximal Self-Association.
Cell Rep., 8, 2014
|
|
8HIK
 
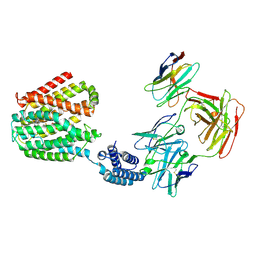 | | The TPP-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HII
 
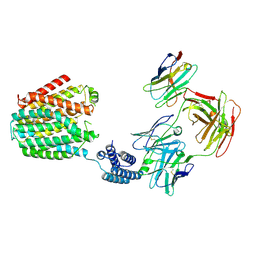 | | The BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | BRIL-SLC19A1 chimera, anti-BRIL Fab heavy chain, anti-BRIL Fab light chain, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
7MPZ
 
 | |
4V2P
 
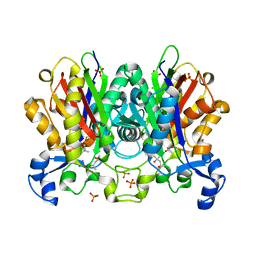 | | Ketosynthase MxnB | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, KETOSYNTHASE, ... | | Authors: | Koehnke, J. | | Deposit date: | 2014-10-13 | | Release date: | 2015-08-26 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | In vitro reconstitution of alpha-pyrone ring formation in myxopyronin biosynthesis.
Chem Sci, 6, 2015
|
|
6DZ7
 
 | |
6E1O
 
 | |
6E0H
 
 | |
2ASN
 
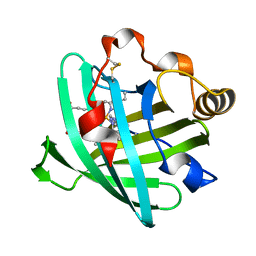 | | Crystal structure of D1A mutant of nitrophorin 2 complexed with imidazole | | Descriptor: | IMIDAZOLE, Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Berry, R.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 2005-08-23 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus
To be Published
|
|
7P4U
 
 | |
2AV7
 
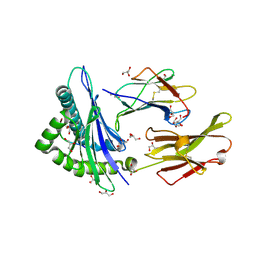 | |
2AH7
 
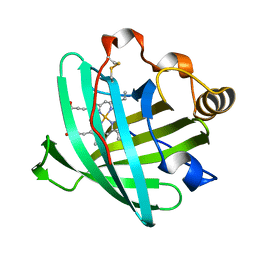 | | Crystal structure of nitrophorin 2 aqua complex | | Descriptor: | Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Berry, R.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 2005-07-27 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus
To be Published
|
|
7MLB
 
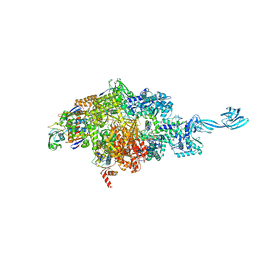 | |
7MLI
 
 | |
7P85
 
 | |
7MLJ
 
 | |
4URR
 
 | | Tailspike protein of Sf6 bacteriophage bound to Shigella flexneri O- antigen octasaccharide fragment | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL TAIL PROTEIN, MANGANESE (II) ION, ... | | Authors: | Gohlke, U, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2014-07-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bacteriophage Tailspikes and Bacterial O-Antigens as a Model System to Study Weak-Affinity Protein-Polysaccharide Interactions.
J.Am.Chem.Soc., 138, 2016
|
|
7M23
 
 | | Human carbonic anhydrase II in complex with troglitazone | | Descriptor: | (5R)-5-(4-{[(2R)-6-HYDROXY-2,5,7,8-TETRAMETHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL]METHOXY}BENZYL)-1,3-THIAZOLIDINE-2,4-DIONE, Carbonic anhydrase 2, ZINC ION | | Authors: | Mueller, S.L, Peat, T.S. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Glitazone Class of Drugs as Carbonic Anhydrase Inhibitors-A Spin-Off Discovery from Fragment Screening.
Molecules, 26, 2021
|
|
7M26
 
 | | Human carbonic anhydrase II in complex with pioglitazone | | Descriptor: | (5R)-5-{4-[2-(5-ethylpyridin-2-yl)ethoxy]benzyl}-1,3-thiazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Mueller, S.L, Peat, T.S. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Glitazone Class of Drugs as Carbonic Anhydrase Inhibitors-A Spin-Off Discovery from Fragment Screening.
Molecules, 26, 2021
|
|
8GYH
 
 | | Crystal structure of Fic25 (apo form) from Streptomyces ficellus | | Descriptor: | DegT/DnrJ/EryC1/StrS family aminotransferase, GLYCEROL, IMIDAZOLE | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-09-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of Sugar Aminotransferase-like Enzymes to Synthesize Stereoisomers of Non-proteinogenic Amino Acids in Natural Product Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
