8BPX
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete composition) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-11-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BJT
 
 | | Structure of human PLK1 in complex with 2-Allyl-1-[6-(1-hydroxy-1-methyl-ethyl)-pyridin-2-yl]-6-[4-(4-methyl-piperazin-1-yl)-phenylamino]-1,2-dihydro-pyrazolo[3,4-d]pyrimidin-3-one | | Descriptor: | 1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, ACETATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Musil, D, Liu-Bujalski, L. | | Deposit date: | 2022-11-06 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.188 Å) | | Cite: | Selective Wee1 Inhibitors Led to Antitumor Activity In Vitro and Correlated with Myelosuppression.
Acs Med.Chem.Lett., 14, 2023
|
|
8BS9
 
 | | Structure of USP36 in complex with Ubiquitin-PA | | Descriptor: | Polyubiquitin-B, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase 36, ... | | Authors: | O'Dea, R, Gersch, M. | | Deposit date: | 2022-11-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for ubiquitin/Fubi cross-reactivity in USP16 and USP36.
Nat.Chem.Biol., 19, 2023
|
|
8BS3
 
 | | Structure of USP36 in complex with Fubi-PA | | Descriptor: | 40S ribosomal protein S30, Ubiquitin carboxyl-terminal hydrolase 36, ZINC ION, ... | | Authors: | O'Dea, R, Gersch, M. | | Deposit date: | 2022-11-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for ubiquitin/Fubi cross-reactivity in USP16 and USP36.
Nat.Chem.Biol., 19, 2023
|
|
8BDE
 
 | | Tubulin-baccatin III complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Lucena-Agell, D, Ma, Y, Estevez-Gallego, J, Li, S, Bargsten, K, Altmann, K.H, Gaillard, N, Kamimura, S, Muehlethaler, T, Gago, F, Oliva, M.A, Steinmetz, M.O, Fang, W.S, Diaz, J.F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural insight into the stabilization of microtubules by taxanes.
Elife, 12, 2023
|
|
8BDG
 
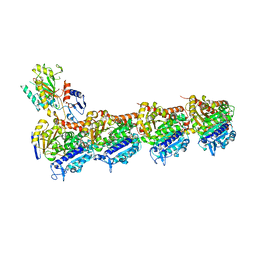 | | Tubulin-taxane-2b complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Lucena-Agell, D, Ma, Y, Estevez-Gallego, J, Li, S, Bargsten, K, Altmann, K.H, Gaillard, N, Kamimura, S, Muehlethaler, T, Gago, F, Oliva, M.A, Steinmetz, M.O, Fang, W.S, Diaz, J.F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insight into the stabilization of microtubules by taxanes.
Elife, 12, 2023
|
|
8BDF
 
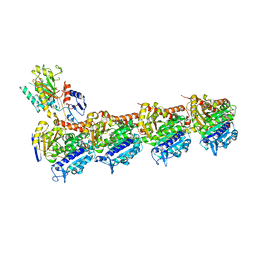 | | Tubulin-taxane-2a complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Lucena-Agell, D, Ma, Y, Estevez-Gallego, J, Li, S, Bargsten, K, Altmann, K.H, Gaillard, N, Kamimura, S, Muehlethaler, T, Gago, F, Oliva, M.A, Steinmetz, M.O, Fang, W.S, Diaz, J.F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight into the stabilization of microtubules by taxanes.
Elife, 12, 2023
|
|
8AY9
 
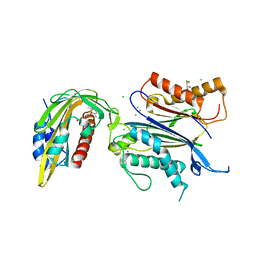 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-ABA-HAB1 TERNARY COMPLEX | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL1, CHLORIDE ION, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
8BAP
 
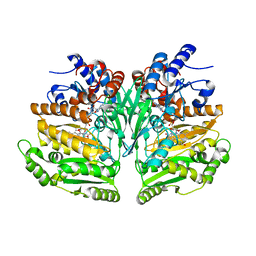 | | Eugenol Oxidase (EUGO) from Rhodococcus jostii RHA1, eightfold mutant active on propanol syringol | | Descriptor: | 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2,6-dimethoxyphenol, CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alvigini, L, Mattevi, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | One-Pot Biocatalytic Synthesis of rac -Syringaresinol from a Lignin-Derived Phenol.
Acs Catalysis, 13, 2023
|
|
7ZX2
 
 | | Tubulin-Pelophen B complex | | Descriptor: | (3R,4S,7S,9S,11S)-3,4,11-trihydroxy-7-((R,Z)-4-(hydroxymethyl)hex-2-en-2-yl)-9-methoxy-12,12-dimethyl-6-oxa-1(1,3)-benzenacyclododecaphan-5-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Estevez-Gallego, J, Diaz, J.F, Van der Eycken, J, Oliva, M.A. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical modulation of microtubule structure through the laulimalide/peloruside site.
Structure, 31, 2023
|
|
8BCV
 
 | | Photosystem I assembly intermediate of Avena sativa | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A, Nelson, N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Photosystem I assembly intermediate of Avena sativa
To Be Published
|
|
8BOW
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with an inhibitor 617 | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[[(2~{S})-3-naphthalen-2-yl-2-[[4-[[2-[(11~{Z})-4,7,10-tris(2-hydroxy-2-oxoethyl)-1,4,7,10-tetrazacyclododec-11-en-1-yl]ethanoylamino]methyl]cyclohexyl]carbonylamino]propanoyl]amino]-1-oxidanyl-1-oxidanylidene-hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Motlova, L, Barinka, C, Benesova, M. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with an inhibitor 617
To Be Published
|
|
1CF9
 
 | |
1QF7
 
 | |
5LWO
 
 | | Structure of Spin-labelled T4 lysozyme mutant L115C-R119C-R1 at 100K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Loll, B, Consentius, P, Gohlke, U, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2016-09-18 | | Release date: | 2017-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Internal Dynamics of the 3-Pyrroline-N-Oxide Ring in Spin-Labeled Proteins.
J Phys Chem Lett, 8, 2017
|
|
5EE7
 
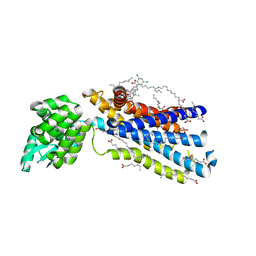 | | Crystal structure of the human glucagon receptor (GCGR) in complex with the antagonist MK-0893 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3-[[4-[(1~{S})-1-[3-[3,5-bis(chloranyl)phenyl]-5-(6-methoxynaphthalen-2-yl)pyrazol-1-yl]ethyl]phenyl]carbonylamino]propanoic acid, Glucagon receptor,Endolysin,Glucagon receptor, ... | | Authors: | Jazayeri, A, Dore, A.S, Lamb, D, Krishnamurthy, H, Southall, S.M, Baig, A.H, Bortolato, A, Koglin, M, Robertson, N.J, Errey, J.C, Andrews, S.P, Brown, A.J.H, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extra-helical binding site of a glucagon receptor antagonist.
Nature, 533, 2016
|
|
2QB0
 
 | |
4W55
 
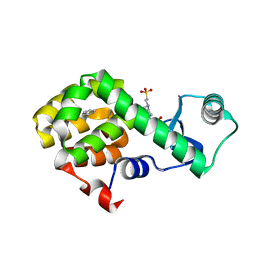 | | T4 Lysozyme L99A with n-Propylbenzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, propylbenzene | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6401 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W58
 
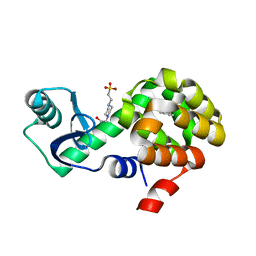 | | T4 Lysozyme L99A with n-Pentylbenzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, pentylbenzene | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7P2L
 
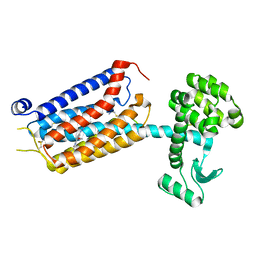 | | thermostabilised 7TM domain of human mGlu5 receptor bound to photoswitchable ligand alloswitch-1 | | Descriptor: | 2-chloranyl-~{N}-[2-methoxy-4-[(~{E})-pyridin-2-yldiazenyl]phenyl]benzamide, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5 | | Authors: | Huang, C.Y, Vinothkumar, K.R, Lebon, G. | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Agonists and allosteric modulators promote signaling from different metabotropic glutamate receptor 5 conformations.
Cell Rep, 36, 2021
|
|
6FFI
 
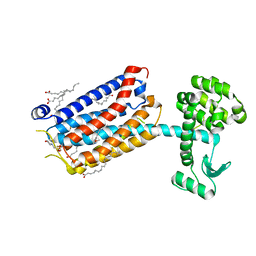 | | Crystal Structure of mGluR5 in complex with MMPEP at 2.2 A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-(3-methoxyphenyl)ethynyl]-6-methyl-pyridine, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | Authors: | Christopher, J.A, Orgovan, Z, Congreve, M, Dore, A.S, Errey, J.C, Marshall, F.H, Mason, J.S, Okrasa, K, Rucktooa, P, Serrano-Vega, M.J, Ferenczy, G.G, Keseru, G.M. | | Deposit date: | 2018-01-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures.
J.Med.Chem., 62, 2019
|
|
6FFH
 
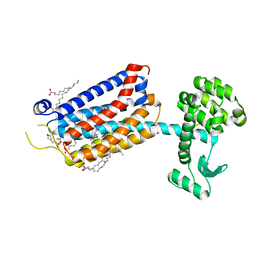 | | Crystal Structure of mGluR5 in complex with Fenobam at 2.65 A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(3-chlorophenyl)-3-(3-methyl-5-oxidanylidene-4~{H}-imidazol-2-yl)urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Christopher, J.A, Orgovan, Z, Congreve, M, Dore, A.S, Errey, J.C, Marshall, F.H, Mason, J.S, Okrasa, K, Rucktooa, P, Serrano-Vega, M.J, Ferenczy, G.G, Keseru, G.M. | | Deposit date: | 2018-01-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures.
J.Med.Chem., 62, 2019
|
|
5EWX
 
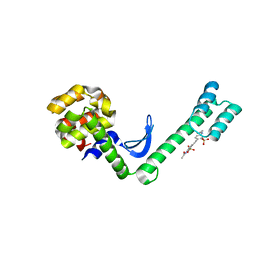 | | Fusion protein of T4 lysozyme and B4 domain of protein A from staphylococcal aureus with chemical cross-linker EY-CBS | | Descriptor: | 2,2'-ethyne-1,2-diylbis{5-[(chloroacetyl)amino]benzenesulfonic acid}, Endolysin,Immunoglobulin G-binding protein A,Endolysin | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-11-22 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
5G27
 
 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at Room Temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, ENDOLYSIN, ... | | Authors: | Gohlke, U, Consentius, P, Loll, B, Mueller, R, Kaupp, M, Heinemann, U, Risse, T. | | Deposit date: | 2016-04-07 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-Ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
6IIH
 
 | | crystal structure of mitochondrial calcium uptake 2(MICU2) | | Descriptor: | CALCIUM ION, Endolysin,Calcium uptake protein 2, mitochondrial | | Authors: | Shen, Q, Wu, W, Zheng, J, Jia, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | The crystal structure of MICU2 provides insight into Ca2+binding and MICU1-MICU2 heterodimer formation.
Embo Rep., 20, 2019
|
|
