2E2J
 
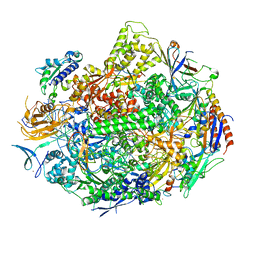 | | RNA polymerase II elongation complex in 5 mM Mg+2 with GMPCPP | | Descriptor: | 27-MER DNA template strand, 5'-D(P*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*A)-3', 5'-R(P*AP*UP*CP*GP*AP*GP*AP*GP*G)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
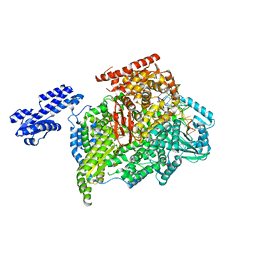
8BYQ
 
 | | RNA polymerase II pre-initiation complex with the proximal +1 nucleosome (PIC-Nuc10W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
5AMR
 
 | |
8BZ1
 
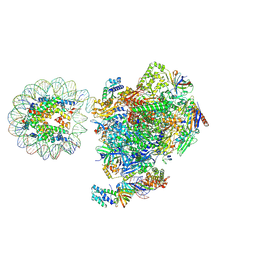 | | RNA polymerase II core pre-initiation complex with the proximal +1 nucleosome (cPIC-Nuc10W) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
6E53
 
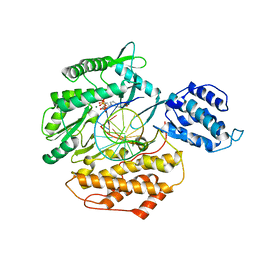 | | Structure of TERT in complex with a novel telomerase inhibitor | | Descriptor: | MAGNESIUM ION, RNA/DNA hairpin, Telomerase reverse transcriptase, ... | | Authors: | Hernandez-Sanchez, W, Huang, W, Plucinsky, B, Garcia-Vazquez, N, Berdis, A.J, Skordalakes, E, Taylor, D.J. | | Deposit date: | 2018-07-19 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A non-natural nucleotide uses a specific pocket to selectively inhibit telomerase activity.
Plos Biol., 17, 2019
|
|
8C4S
 
 | |
8C2P
 
 | | FMDV 3D polymerase in complex with 3B3 | | Descriptor: | Protein 3B-3, RNA-directed RNA polymerase 3D-POL | | Authors: | Ferrer-Orta, C, Veraguer, N. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dual role of the foot-and-mouth disease virus 3B1 protein in the replication complex: As protein primer and as an essential component to recruit 3Dpol to membranes.
Plos Pathog., 19, 2023
|
|
8C1N
 
 | |
8BVW
 
 | | RNA polymerase II pre-initiation complex with the distal +1 nucleosome (PIC-Nuc18W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
8G6J
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2) | | Descriptor: | (3R,6R,9S,12S,15S,18S,20R,24aR)-6-[(2S)-butan-2-yl]-3,12-bis[(1R)-1-hydroxy-2-methylpropyl]-8,9,11,17,18-pentamethyl-15-[(2S)-2-methylbutyl]hexadecahydropyrido[1,2-a][1,4,7,10,13,16,19]heptaazacyclohenicosine-1,4,7,10,13,16,19(21H)-heptone, (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 1,4-DIAMINOBUTANE, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G60
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Z
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G61
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Y
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8GLP
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Watson, Z.L, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8CI5
 
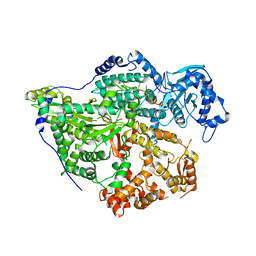 | | Structure of the SNV L protein bound to 5' RNA | | Descriptor: | RNA (5'-R(P*AP*GP*UP*AP*GP*UP*AP*GP*AP*CP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Meier, K, Thorkelsson, S.R, Durieux Trouilleton, Q, Vogel, D, Yu, D, Kosinski, J, Cusack, S, Malet, H, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-02-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of the Sin Nombre virus L protein.
Plos Pathog., 19, 2023
|
|
7Z1L
 
 | | Structure of yeast RNA Polymerase III Pre-Termination Complex (PTC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1N
 
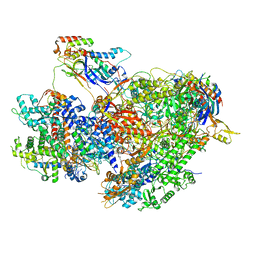 | | Structure of yeast RNA Polymerase III Delta C53-C37-C11 | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
6T0W
 
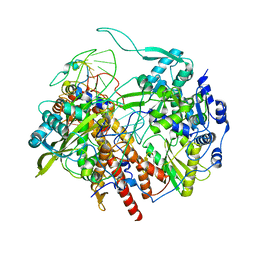 | | Human Influenza B polymerase recycling complex | | Descriptor: | Polymerase PB2, Polymerase acidic protein, RNA-directed RNA polymerase catalytic subunit, ... | | Authors: | Wandzik, J.M, Kouba, T, Karuppasamy, M, Cusack, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
6T0U
 
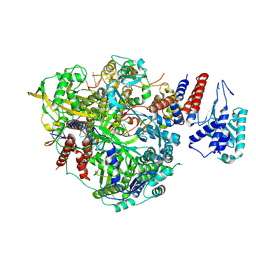 | | Bat Influenza A polymerase product dissociation complex using 44-mer vRNA template with intact oligo(U) sequence | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit, ... | | Authors: | Wandzik, J.M, Kouba, T, Cusack, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
6T0R
 
 | | Bat Influenza A polymerase product dissociation complex using 44-mer vRNA template with mutated oligo(U) sequence | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit, ... | | Authors: | Wandzik, J.M, Kouba, T, Cusack, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
6GZ3
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-1 (TI-POST-1) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
8JSM
 
 | | The structure of EBOV L-VP35-RNA complex (conformation 1) | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV genome., ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
8JSL
 
 | | The structure of EBOV L-VP35-RNA complex | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV, ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
8JSN
 
 | | The structure of EBOV L-VP35-RNA complex (conformation 2) | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV genome, ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
