5NBV
 
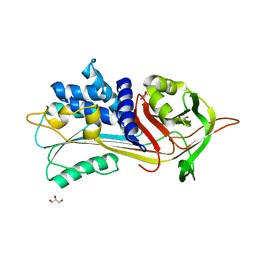 | |
5NB7
 
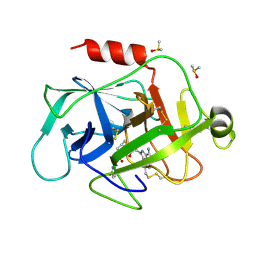 | | Complement factor D | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[(6-bromanylpyridin-2-yl)carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]indazole-3-carboxamide, Complement factor D, DIMETHYL SULFOXIDE | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NOG
 
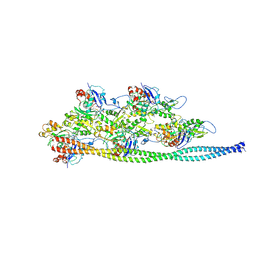 | | Ca2+-induced Movement of Tropomyosin on Native Cardiac Thin Filaments - "Blocked" state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cardiac muscle alpha actin 1, MAGNESIUM ION, ... | | Authors: | Risi, C, Eisner, J, Belknap, B, Heeley, D.H, White, H.D, Schroeder, G.F, Galkin, V.E. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-19 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Ca(2+)-induced movement of tropomyosin on native cardiac thin filaments revealed by cryoelectron microscopy.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NP3
 
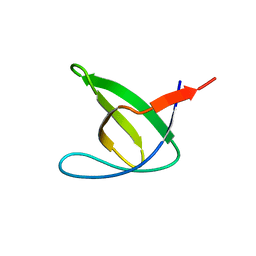 | | Abl2 SH3 | | Descriptor: | Abelson tyrosine-protein kinase 2 | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
5DDO
 
 | |
8J4K
 
 | |
5DGX
 
 | | 1.73 Angstrom resolution crystal structure of the ABC-ATPase domain (residues 357-609) of lipid A transport protein (msbA) from Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1.73 Angstrom resolution crystal structure of the ABC-ATPase domain (residues 357-609) of lipid A transport protein (msbA) from Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP
To Be Published
|
|
8IZC
 
 | | Human CK1 Delta Kinase structure bound to Inhibitor | | Descriptor: | Casein kinase I isoform delta, SULFATE ION, ~{N}5-~{tert}-butyl-2-(3-chloranyl-4-fluoranyl-phenyl)-6,7-dihydro-4~{H}-pyrazolo[1,5-a]pyrazine-3,5-dicarboxamide | | Authors: | Ghosh, K. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Discovery of BMS-986164, a Potent, Selective and Orally Efficacious CK1d/e/a Inhibitor from Pyrazolo-Piperazine Chemotype
To Be Published
|
|
5N71
 
 | | CRYSTAL STRUCTURE OF MATURE CATHEPSIN D FROM THE TICK IXODES RICINUS (IRCD1) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative cathepsin d, SULFATE ION | | Authors: | Brynda, J, Hanova, I, Hobizalova, R, Mares, M. | | Deposit date: | 2017-02-17 | | Release date: | 2017-12-27 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Novel Structural Mechanism of Allosteric Regulation of Aspartic Peptidases via an Evolutionarily Conserved Exosite.
Cell Chem Biol, 25, 2018
|
|
5N7Q
 
 | | CRYSTAL STRUCTURE OF MATURE CATHEPSIN D FROM THE TICK IXODES RICINUS (IRCD1) IN COMPLEX WITH THE INHIBITOR PEPSTATIN A | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PEPSTATIN A, ... | | Authors: | Brynda, J, Hanova, I, Hobizalova, R, Mares, M. | | Deposit date: | 2017-02-21 | | Release date: | 2017-12-27 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Structural Mechanism of Allosteric Regulation of Aspartic Peptidases via an Evolutionarily Conserved Exosite.
Cell Chem Biol, 25, 2018
|
|
5CPP
 
 | |
5D3U
 
 | | Crystal structure of the 5-selective H176F mutant of Cytochrome TxtE | | Descriptor: | CHLORIDE ION, GLYCEROL, P450-like protein, ... | | Authors: | Cahn, J.K.B, Dodani, S.C, Arnold, F.H. | | Deposit date: | 2015-08-06 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of a regioselectivity switch in nitrating P450s guided by molecular dynamics simulations and Markov models.
Nat.Chem., 8, 2016
|
|
5N2N
 
 | | Crystal structure of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana complexed with Mg2+ and BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Histidine kinase CKI1, MAGNESIUM ION | | Authors: | Otrusinova, O, Demo, G, Padrta, P, Jasenakova, Z, Pekarova, B, Gelova, Z, Szmitkowska, A, Kaderavek, P, Jansen, S, Zachrdla, M, Klumpler, T, Marek, J, Hritz, J, Janda, L, Iwai, H, Wimmerova, M, Hejatko, J, Zidek, L. | | Deposit date: | 2017-02-08 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational dynamics are a key factor in signaling mediated by the receiver domain of a sensor histidine kinase from Arabidopsis thaliana.
J. Biol. Chem., 292, 2017
|
|
8JG8
 
 | |
5NE0
 
 | | Room temperature in-situ structure of hen egg-white lysozyme from crystals enclosed between ultrathin silicon nitride membranes | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Martiel, I, Opara, N, Arnold, S.A, Braun, T, Stahlberg, H, Makita, M, David, C, Padeste, C. | | Deposit date: | 2017-03-09 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Direct protein crystallization on ultrathin membranes for diffraction measurements at X-ray free-electron lasers.
J.Appl.Crystallogr., 50, 2017
|
|
5DE9
 
 | | The role of Ile87 of CYP158A2 in oxidative coupling reaction | | Descriptor: | Biflaviolin synthase CYP158A2, PROTOPORPHYRIN IX CONTAINING FE, SPERMINE | | Authors: | Zhao, B. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The role of Ile87 of CYP158A2 in oxidative coupling reaction.
Arch. Biochem. Biophys., 518, 2012
|
|
5N14
 
 | |
5D5C
 
 | | In meso in situ serial X-ray crystallography structure of lysozyme at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D97
 
 | | Neutron crystal structure of H2O-solvent ribonuclease A | | Descriptor: | ISOPROPYL ALCOHOL, Ribonuclease pancreatic | | Authors: | Chatake, T, Fujiwara, S. | | Deposit date: | 2015-08-18 | | Release date: | 2016-04-06 | | Last modified: | 2018-12-05 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | A technique for determining the deuterium/hydrogen contrast map in neutron macromolecular crystallography
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8IYG
 
 | | Human neuronal gap junction channel connexin 36 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, Gap junction delta-2 protein | | Authors: | Mao, W.X, Chen, S.S. | | Deposit date: | 2023-04-04 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Assembly mechanisms of the neuronal gap junction channel connexin 36 elucidated by Cryo-EM.
Arch.Biochem.Biophys., 754, 2024
|
|
5NAR
 
 | |
5AHA
 
 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-(2-methoxyethoxy)hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[(2Z)-2-(methylimino)-2,3-dihydro-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5NAW
 
 | | Complement factor D in complex with the inhibitor (1R,3S,5R)-2-Aza-bicyclo[3.1.0]hexane-2,3-dicarboxylic acid 2-[(1-carbamoyl-1H-indol-3-yl)-amide] 3-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (1~{R},3~{S},5~{R})-~{N}2-(1-aminocarbonylindol-3-yl)-~{N}3-[3-(trifluoromethyloxy)phenyl]-2-azabicyclo[3.1.0]hexane-2,3-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-02-28 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NBL
 
 | | Crystal structure of the Arp4-N-actin(APO-state) heterodimer bound by a nanobody | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Knoll, K.R, Eustermann, S, Hopfner, K.P. | | Deposit date: | 2017-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5A5J
 
 | | Cytochrome 2C9 P450 inhibitor complex | | Descriptor: | CYTOCHROME P450 2C9, N-[4-(3-chloranyl-4-cyano-phenoxy)-3,5-dimethoxy-phenyl]-1,1,1-tris(fluoranyl)methanesulfonamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Skerratt, S.E, de Groot, M.J, Phillips, C. | | Deposit date: | 2015-06-18 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a Novel Binding Pocket for Cyp 2C9 Inhibitors: Crystallography, Pharmacophore Modelling and Inhibitor Sar.
To be Published
|
|
