3L15
 
 | | Human Tead2 transcriptional factor | | Descriptor: | GLYCEROL, Transcriptional enhancer factor TEF-4 | | Authors: | Tomchick, D.R, Luo, X, Tian, W. | | Deposit date: | 2009-12-10 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the YAP-binding domain of human TEAD2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KM5
 
 | | Crystal Structure Analysis of the K2 Cleaved Adhesin Domain of Lys-gingipain (Kgp) | | Descriptor: | CALCIUM ION, GLYCEROL, Lysine specific cysteine protease, ... | | Authors: | Li, N, Collyer, C.A, Hunter, N. | | Deposit date: | 2009-11-09 | | Release date: | 2010-03-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure determination and analysis of a haemolytic gingipain adhesin domain from Porphyromonas gingivalis
Mol.Microbiol., 76, 2010
|
|
3L22
 
 | |
3KMG
 
 | |
3L2J
 
 | |
3KN7
 
 | |
3KNC
 
 | |
3KNP
 
 | |
3KNZ
 
 | |
3KBZ
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 6 | | Descriptor: | Fructose-1,6-bisphosphatase 1, {[(2-amino-8H-indeno[1,2-d][1,3]thiazol-4-yl)oxy]methyl}phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KDU
 
 | |
3KDW
 
 | |
3KE5
 
 | |
3KF5
 
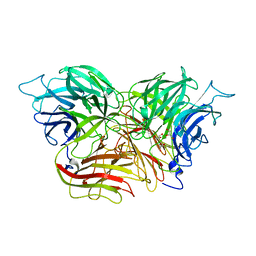 | | Structure of invertase from Schwanniomyces occidentalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Invertase | | Authors: | Sanz-Aparicio, J, Polo, A. | | Deposit date: | 2009-10-27 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and kinetic analysis of Schwanniomyces occidentalis invertase reveals a new oligomerization pattern and the role of its supplementary domain in substrate binding
J.Biol.Chem., 285, 2010
|
|
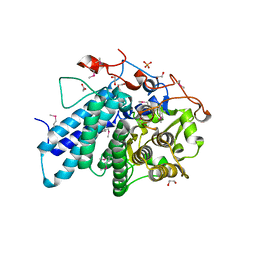
3KEZ
 
 | |
3KC1
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 19a | | Descriptor: | Fructose-1,6-bisphosphatase 1, {[(7-carbamoyl-8H-indeno[1,2-d][1,3]thiazol-4-yl)oxy]methyl}phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KFN
 
 | |
3KCT
 
 | | CRYSTAL STRUCTURE OF PAmCherry1 in the photoactivated state | | Descriptor: | PAmCherry1 protein | | Authors: | Malashkevich, V.N, Subach, F.V, Zencheck, W.D, Xiao, H, Filonov, G.S, Almo, S.C, Verkhusha, V.V. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3KDT
 
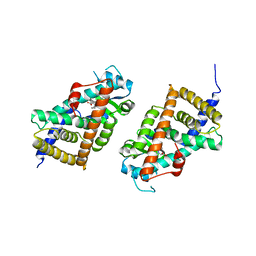 | |
3KGU
 
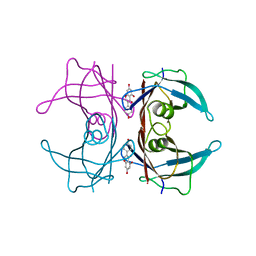 | |
3KE3
 
 | |
3KH1
 
 | |
3KEX
 
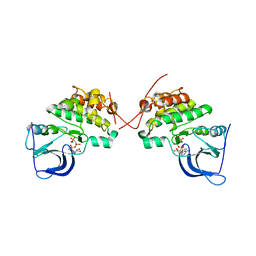 | | Crystal structure of the catalytically inactive kinase domain of the human epidermal growth factor receptor 3 (HER3) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Jura, N, Shan, Y, Cao, X, Shaw, D.E, Kuriyan, J. | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3KJ2
 
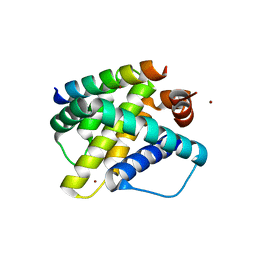 | | Mcl-1 in complex with Bim BH3 mutant F4aE | | Descriptor: | ACETATE ION, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3KJH
 
 | | Zn-bound state of CooC1 | | Descriptor: | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC, ZINC ION | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2009-11-03 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the ATP-Dependent Maturation Factor of Ni,Fe-Containing Carbon Monoxide Dehydrogenases
J.Mol.Biol., 396, 2010
|
|
