8BSC
 
 | |
5E01
 
 | | Crystal structure of HiNmlR, a MerR family regulator lacking the sensor domain, bound to palyndromic promoter DNA | | Descriptor: | 5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*GP*CP*AP*CP*TP*CP*TP*AP*AP*G)-3', Uncharacterized HTH-type transcriptional regulator HI_0186 | | Authors: | Counago, R.M, Chang, C.W, Chen, N.H, Djoko, K.Y, McEwan, A.G, Kobe, B. | | Deposit date: | 2015-09-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of thiol-based regulation of formaldehyde detoxification in H. influenzae by a MerR regulator with no sensor region.
Nucleic Acids Res., 44, 2016
|
|
2GIH
 
 | | Q138F HincII bound to cognate DNA GTCGAC and Ca2+ | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*CP*GP*AP*CP*CP*GP*GP*C)-3', CALCIUM ION, Type II restriction enzyme HincII | | Authors: | Horton, N.C, Joshi, H.K, Etzkorn, C, Chatwell, L, Bitinaite, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
J.Biol.Chem., 281, 2006
|
|
3KHR
 
 | | Dpo4 post-extension ternary complex with the correct C opposite the 2-aminofluorene-guanine [AF]G lesion | | Descriptor: | 2-AMINOFLUORENE, 5'-D(*CP*C*TP*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*CP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5VU7
 
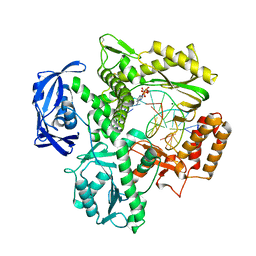 | | TNA polymerase, open ternary complex | | Descriptor: | 5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXYTETRAHYDROFURAN-3-YL DIHYDROGEN PHOSPHATE, DNA polymerase, DNA template, ... | | Authors: | Chim, N, Chaput, J.C. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-06 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis for TNA synthesis by an engineered TNA polymerase.
Nat Commun, 8, 2017
|
|
4F1I
 
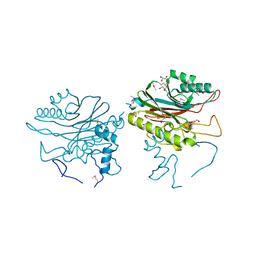 | |
3U3W
 
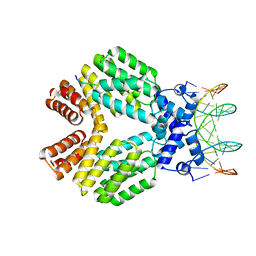 | | Crystal Structure of Bacillus thuringiensis PlcR in complex with the peptide PapR7 and DNA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*AP*AP*AP*TP*AP*TP*TP*GP*CP*AP*TP*AP*G)-3', 5'-D(P*CP*TP*AP*TP*GP*CP*AP*AP*TP*AP*TP*TP*TP*CP*AP*TP*AP*T)-3', C-terminus heptapeptide from PapR protein, ... | | Authors: | Grenha, R, Slamti, L, Bouillaut, L, Lereclus, D, Nessler, S. | | Deposit date: | 2011-10-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8GLX
 
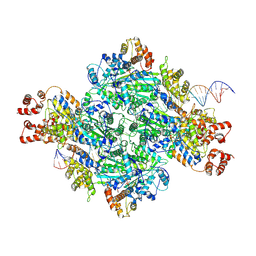 | |
2XWC
 
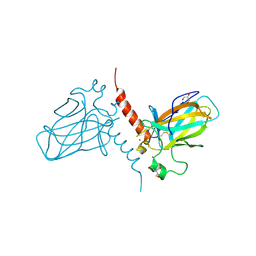 | | Crystal structure of the DNA binding domain of human TP73 refined at 1.8 A resolution | | Descriptor: | GLYCEROL, TRIS(HYDROXYETHYL)AMINOMETHANE, TUMOUR PROTEIN P73, ... | | Authors: | Canning, P, Zhang, Y, Vollmar, M, Krojer, T, Ugochukwu, E, Muniz, J.R.C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Aspp2 Recognition by the Tumor Suppressor P73.
J.Mol.Biol., 423, 2012
|
|
8GLW
 
 | |
6R4S
 
 | |
7CVQ
 
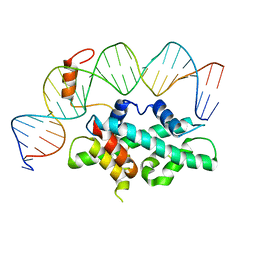 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB2/YC3 and FT CORE1 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-3 and Zinc finger protein CONSTANS, FT CORE1 DNA forward strand, FT CORE1 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
7CVO
 
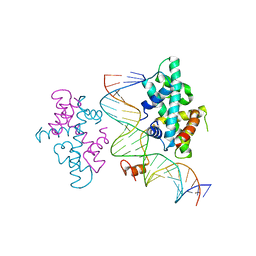 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB3/YC4 and FT CORE2 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-4 and Zinc finger protein CONSTANS, FT CORE2 DNA forward strand, FT CORE2 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
6R4T
 
 | |
1VQB
 
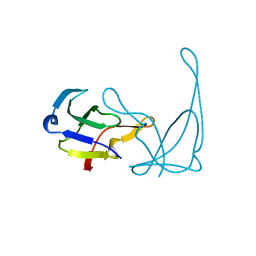 | | GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Skinner, M.M, Zhang, H, Leschnitzer, D.H, Guan, Y, Bellamy, H, Sweet, R.M, Gray, C.W, Konings, R.N.H, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the gene V protein of bacteriophage f1 determined by multiwavelength x-ray diffraction on the selenomethionyl protein.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
6R5E
 
 | |
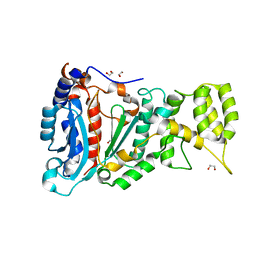
6RB4
 
 | |
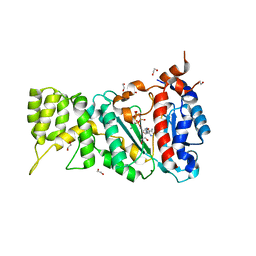
6R5D
 
 | |
6R4U
 
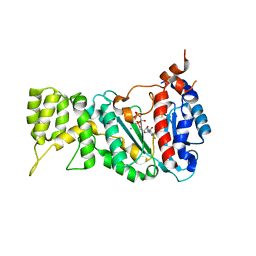 | |
4FB3
 
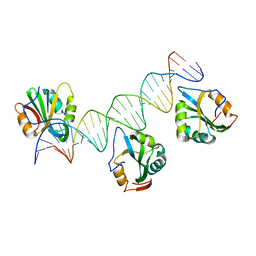 | | Polyomavirus T-ag binds symmetrical repeats at the viral origin in an asymmetrical manner | | Descriptor: | Large T antigen, ORI DNA oligonucleotide-Crick strand, ORI DNA oligonucleotide-Watson strand | | Authors: | Bohm, A, Harrison, C.J, Schaffhausen, B.S, Jiang, T. | | Deposit date: | 2012-05-22 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Polyomavirus large T antigen binds symmetrical repeats at the viral origin in an asymmetrical manner.
J.Virol., 87, 2013
|
|
1ID3
 
 | | CRYSTAL STRUCTURE OF THE YEAST NUCLEOSOME CORE PARTICLE REVEALS FUNDAMENTAL DIFFERENCES IN INTER-NUCLEOSOME INTERACTIONS | | Descriptor: | HISTONE H2A.1, HISTONE H2B.2, HISTONE H3, ... | | Authors: | White, C.L, Suto, R.K, Luger, K. | | Deposit date: | 2001-04-03 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions.
EMBO J., 20, 2001
|
|
5LAR
 
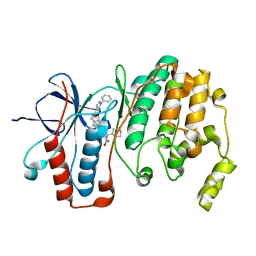 | | Crystal structure of p38 alpha MAPK14 in complex with VPC00628 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Petersen, L.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel p38alpha MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library
Medchemcomm, 7, 2016
|
|
4OB4
 
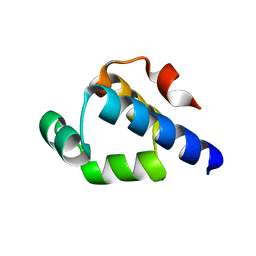 | | Structure of the S. venezulae BldD DNA-binding domain | | Descriptor: | Putative DNA-binding protein | | Authors: | schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
1HNI
 
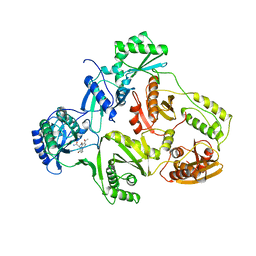 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN A COMPLEX WITH THE NONNUCLEOSIDE INHIBITOR ALPHA-APA R 95845 AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Ding, J, Das, K, Arnold, E. | | Deposit date: | 1995-02-28 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase in a complex with the non-nucleoside inhibitor alpha-APA R 95845 at 2.8 A resolution.
Structure, 3, 1995
|
|
7DHY
 
 | | Arsenic-bound p53 DNA-binding domain mutant G245S | | Descriptor: | ARSENIC, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Chen, S, Lu, M. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Arsenic Trioxide Rescues Structural p53 Mutations through a Cryptic Allosteric Site.
Cancer Cell, 39, 2021
|
|
