2MUH
 
 | |
3N2C
 
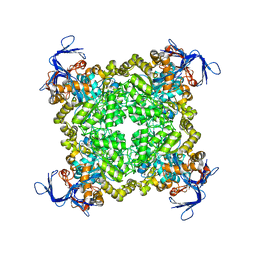 | | Crystal structure of prolidase eah89906 complexed with n-methylphosphonate-l-proline | | Descriptor: | 1-[(R)-hydroxy(methyl)phosphoryl]-L-proline, PROLIDASE, ZINC ION | | Authors: | Patskovsky, Y, Xu, C, Sauder, J.M, Burley, S.K, Raushel, F.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
2N1O
 
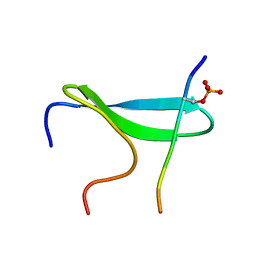 | |
3CZA
 
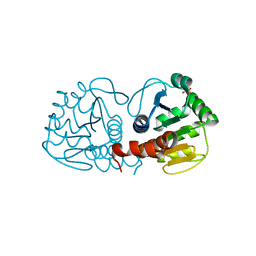 | | Crystal Structure of E18D DJ-1 | | Descriptor: | MALONIC ACID, Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hashim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
2HZP
 
 | | Crystal Structure of Homo Sapiens Kynureninase | | Descriptor: | Kynureninase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lima, S, Khristoforov, R, Momany, C, Phillips, R.S. | | Deposit date: | 2006-08-09 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Homo sapiens Kynureninase.
Biochemistry, 46, 2007
|
|
4IRM
 
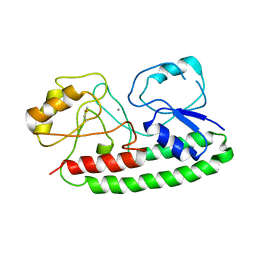 | |
3T95
 
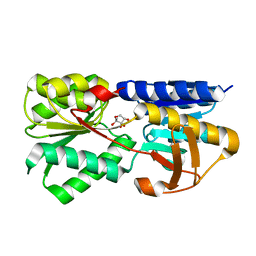 | | Crystal structure of LsrB from Yersinia pestis complexed with autoinducer-2 | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, Autoinducer 2-binding protein lsrB | | Authors: | Kavanaugh, J.S, Gakhar, L, Horswill, A.R. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of LsrB from Yersinia pestis complexed with autoinducer-2.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2YLK
 
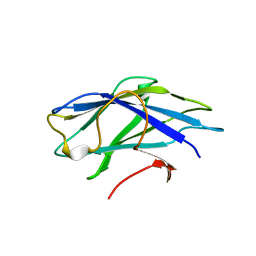 | | Carbohydrate-binding module CBM3b from the cellulosomal cellobiohydrolase 9A from Clostridium thermocellum | | Descriptor: | CELLULOSE 1,4-BETA-CELLOBIOSIDASE | | Authors: | Yaniv, O, Petkun, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2011-06-02 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Single Mutation Reforms the Binding Activity of an Adhesion-Deficient Family 3 Carbohydrate-Binding Module
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3EH9
 
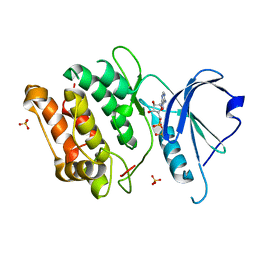 | | Crystal structure of death associated protein kinase complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Death-associated protein kinase 1, SULFATE ION | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
7E0M
 
 | | Crystal structure of phospholipase D | | Descriptor: | Phospholipase, SULFATE ION | | Authors: | Wang, F.H. | | Deposit date: | 2021-01-28 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of a Phospholipase D from the Plant-Associated Bacteria Serratia plymuthica Strain AS9 Reveals a Unique Arrangement of Catalytic Pocket.
Int J Mol Sci, 22, 2021
|
|
4C2J
 
 | | Crystal structure of human mitochondrial 3-ketoacyl-CoA thiolase in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, 3-KETOACYL-COA THIOLASE, MITOCHONDRIAL, ... | | Authors: | Kiema, T.-R, Harijan, R.K, Wierenga, R.K. | | Deposit date: | 2013-08-19 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Mitochondrial 3-Ketoacyl-Coa Thiolase (T1): Insight Into the Reaction Mechanism of its Thiolase and Thioesterase Activities
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2AKQ
 
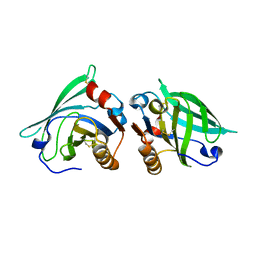 | | The structure of bovine B-lactoglobulin A in crystals grown at very low ionic strength | | Descriptor: | Beta-lactoglobulin variant A | | Authors: | Adams, J.J, Anderson, B.F, Norris, G.E, Creamer, L.K, Jameson, G.B. | | Deposit date: | 2005-08-03 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bovine beta-lactoglobulin (variant A) at very low ionic strength
J.Struct.Biol., 154, 2006
|
|
2L0P
 
 | | Solution structure of human apo-S100A1 protein by NMR spectroscopy | | Descriptor: | S100 calcium binding protein A1 | | Authors: | Nowakowski, M, Jaremko, L, Jaremko, M, Bierzynski, A, Zhukov, I, Ejchart, A. | | Deposit date: | 2010-07-12 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and dynamics of human apo-S100A1 protein.
J.Struct.Biol., 174, 2011
|
|
3CZ9
 
 | | Crystal Structure of E18L DJ-1 | | Descriptor: | O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hasim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
3CYF
 
 | | Crystal Structure of E18N DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hasim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
3IFK
 
 | |
3EHA
 
 | |
4JA1
 
 | | Structure of MMP3 complexed with a platinum-based inhibitor | | Descriptor: | CALCIUM ION, CHLORIDE ION, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Belviso, B.D, Arnesano, F, Calderone, V, Caliandro, R, Natile, G, Siliqi, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of matrix metalloproteinase-3 with a platinum-based inhibitor.
Chem.Commun.(Camb.), 49, 2013
|
|
3EEF
 
 | | Crystal structure of N-carbamoylsarcosine amidase from thermoplasma acidophilum | | Descriptor: | N-carbamoylsarcosine amidase related protein, ZINC ION | | Authors: | Luo, H.-B, Zheng, H, Chruszcz, M, Zimmerman, M.D, Skarina, T, Egorova, O, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and molecular modeling study of N-carbamoylsarcosine amidase Ta0454 from Thermoplasma acidophilum.
J.Struct.Biol., 169, 2010
|
|
2B0F
 
 | |
2JYM
 
 | |
2ZCI
 
 | |
4HL8
 
 | | Re-refinement of the vault ribonucleoprotein particle | | Descriptor: | Major vault protein | | Authors: | Casanas, A, Querol-Audi, J, Guerra, P, Pous, J, Tanaka, H, Tsukihara, T, Verdaguer, V, Fita, I. | | Deposit date: | 2012-10-16 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | New features of vault architecture and dynamics revealed by novel refinement using the deformable elastic network approach.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2MNI
 
 | |
3QL3
 
 | | Re-refined coordinates for PDB entry 1RX2 | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
