2NT8
 
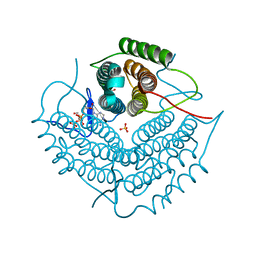 | | ATP bound at the active site of a PduO type ATP:co(I)rrinoid adenosyltransferase from Lactobacillus reuteri | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cobalamin adenosyltransferase, GLYCEROL, ... | | Authors: | St-Maurice, M, Mera, P.E, Taranto, M.P, Sesma, F, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2006-11-07 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural characterization of the active site of the PduO-type ATP:Co(I)rrinoid adenosyltransferase from Lactobacillus reuteri.
J.Biol.Chem., 282, 2007
|
|
3R5H
 
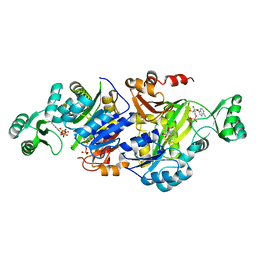 | | Crystal Structure of ADP-AIR complex of purK: N5-carboxyaminoimidazole ribonucleotide synthetase | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fung, L.W, Tuntland, M.L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2011-03-18 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of the bicarbonate binding site and insights into the carboxylation mechanism of (N(5))-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3G32
 
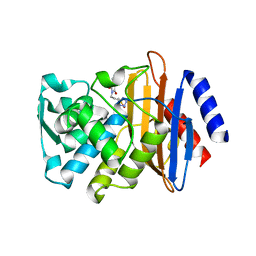 | | CTX-M-9 class A beta-lactamase complexed with compound 6 (3G3) | | Descriptor: | 2-[2-(1H-tetrazol-5-yl)ethyl]-1H-isoindole-1,3(2H)-dione, Beta-lactamase CTX-M-9a, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, Y, Shoichet, B.K. | | Deposit date: | 2009-02-01 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Molecular docking and ligand specificity in fragment-based inhibitor discovery
Nat.Chem.Biol., 5, 2009
|
|
3G31
 
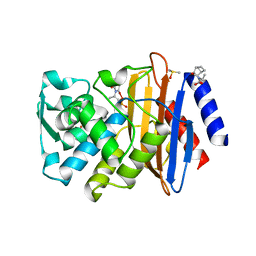 | | CTX-M-9 class A beta-lactamase complexed with compound 4 (GF1) | | Descriptor: | (2S)-2-[(3aR,4R,7S,7aS)-1,3-dioxooctahydro-2H-4,7-methanoisoindol-2-yl]propanoic acid, Beta-lactamase CTX-M-9a, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, Y, Shoichet, B.K. | | Deposit date: | 2009-02-01 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular docking and ligand specificity in fragment-based inhibitor discovery
Nat.Chem.Biol., 5, 2009
|
|
2NTG
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R7 | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(4-bromo-1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-11-07 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
3RMC
 
 | | Crystal Structure of a replicative DNA polymerase bound to DNA containing Thymine Glycol | | Descriptor: | DNA (5'-D(*CP*GP*TP*(CTG)P*GP*AP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*CP*A)-3'), DNA polymerase | | Authors: | Aller, P, Duclos, S, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A crystallographic study of the role of sequence context in thymine glycol bypass by a replicative DNA polymerase serendipitously sheds light on the exonuclease complex.
J.Mol.Biol., 412, 2011
|
|
3G3P
 
 | |
3G3W
 
 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
3R7F
 
 | | Crystal Structure of CP-bound Aspartate Transcarbamoylase from Bacillus subtilis | | Descriptor: | Aspartate carbamoyltransferase, PHOSPHATE ION, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Cockrell, G.M, Harris, K.M, Puleo, D.E, Kantrowitz, E.R. | | Deposit date: | 2011-03-22 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystallographic Snapshots of the Complete Catalytic Cycle of the Unregulated Aspartate Transcarbamoylase from Bacillus subtilis.
J.Mol.Biol., 411, 2011
|
|
3G4S
 
 | | Co-crystal structure of Tiamulin bound to the large ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome.
J.Mol.Biol., 389, 2009
|
|
3R9U
 
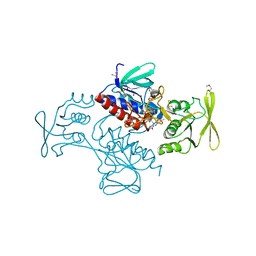 | | Thioredoxin-disulfide reductase from Campylobacter jejuni. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, K.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-25 | | Release date: | 2011-04-06 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Thioredoxin-disulfide reductase from Campylobacter jejuni.
To be Published
|
|
2NY0
 
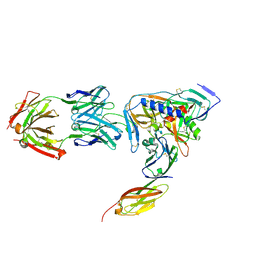 | | HIV-1 gp120 Envelope Glycoprotein (M95W, W96C, T257S, V275C, S334A, S375W, A433M) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
2NZO
 
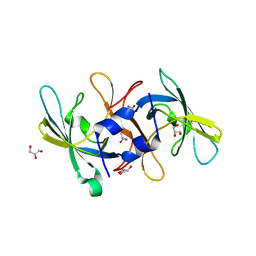 | |
2NYB
 
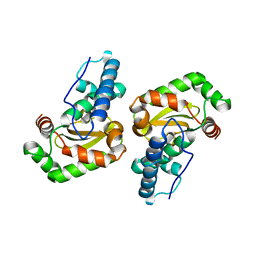 | |
3D9E
 
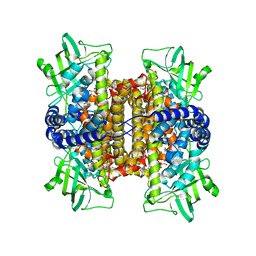 | | Nitroalkane oxidase: active site mutant D402N crystallized with 1-nitrooctane | | Descriptor: | 1-nitrooctane, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
3RC5
 
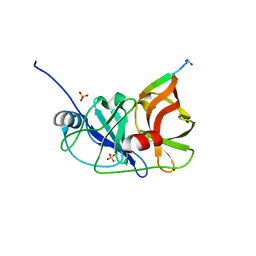 | |
2O0U
 
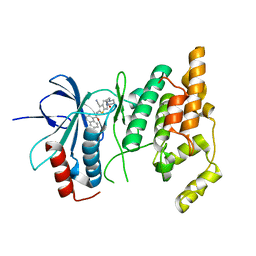 | | Crystal structure of human JNK3 complexed with N-{3-cyano-6-[3-(1-piperidinyl)propanoyl]-4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl}-1-naphthalenecarboxamide | | Descriptor: | Mitogen-activated protein kinase 10, N-{3-CYANO-6-[3-(1-PIPERIDINYL)PROPANOYL]-4,5,6,7-TETRAHYDROTHIENO[2,3-C]PYRIDIN-2-YL}1-NAPHTHALENECARBOXAMIDE | | Authors: | Rowland, P, Somers, D. | | Deposit date: | 2006-11-28 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N-(3-Cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)amides as potent, selective, inhibitors of JNK2 and JNK3.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3DAU
 
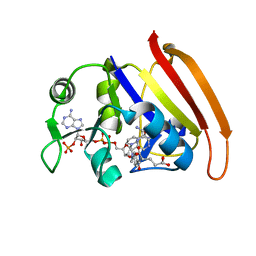 | |
3R0H
 
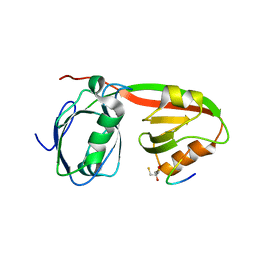 | | Structure of INAD PDZ45 in complex with NG2 peptide | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Inactivation-no-after-potential D protein, ... | | Authors: | Wei, Z, Liu, W, Zhang, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye
Cell(Cambridge,Mass.), 145, 2011
|
|
3DNP
 
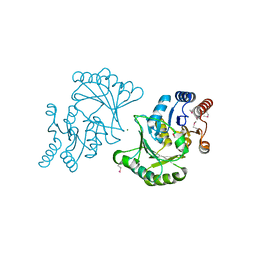 | | Crystal structure of Stress response protein yhaX from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Stress response protein yhaX | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-02 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Stress response protein yhaX from Bacillus subtilis
To be Published
|
|
3DEM
 
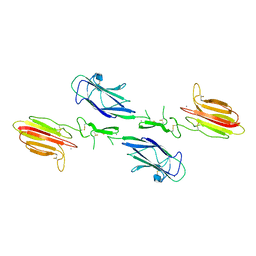 | | CUB1-EGF-CUB2 domain of HUMAN MASP-1/3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement factor MASP-3 | | Authors: | Gaboriaud, C. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the CUB1-EGF-CUB2 domain of human MASP-1/3 and identification of its interaction sites with mannan-binding lectin and ficolins
J.Biol.Chem., 283, 2008
|
|
3R3W
 
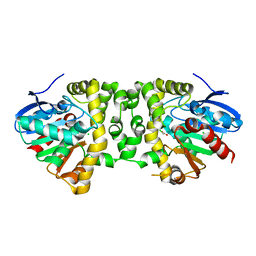 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Chloroacetate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fluoroacetate dehalogenase, ... | | Authors: | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-03-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3DGM
 
 | |
2O3V
 
 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site complexed with paromamine derivative NB33 | | Descriptor: | (2S,3R,4R,5S,6R)-3-AMINO-4-({[(2S,3R,4R,5S,6R)-3-AMINO-2-{[(1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYL]OXY}-5-HYDROXY-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-4-YL]OXY}METHOXY)-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-2,5-DIOL, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
3R5V
 
 | | The structure of calcium bound Thermococcus thioreducens inorganic pyrophosphatase at 298K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Hughes, R.C, Meehan, E.J, Coates, L, Ng, J.D. | | Deposit date: | 2011-03-19 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of calcium bound Thermococcus thioreducens inorganic pyrophosphatase at 298K
To be Published
|
|
