5AQP
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, DIMETHYL SULFOXIDE, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
7VO7
 
 | | Crystal structure of trypsin in complex with Lima bean trypsin inhibitor at 2.25A resolution. | | Descriptor: | Bowman-Birk type proteinase inhibitor, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ahmad, M.S, Akbar, Z, Choudhary, M.I. | | Deposit date: | 2021-10-12 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insight into the structural basis of the dual inhibitory mode of Lima bean (Phaseolus lunatus) serine protease inhibitor.
Proteins, 91, 2023
|
|
6M4P
 
 | | Cytochrome P450 monooxygenase StvP2 substrate-bound structure | | Descriptor: | 6-methoxy-streptovaricin C, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | Deposit date: | 2020-03-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
5AOS
 
 | | Structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A solved at the As edge | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8OLZ
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of K+, Mg2+, 5'-exon, and intronistat B after 2h30 soaking | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-exon, DOMAINS 1-5, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2023-03-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
7TED
 
 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (S,E)-3-amino-4-(fluoromethylene)cyclopent-1-ene-1-carboxylate | | Descriptor: | (1S,3R,4S)-3-formyl-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Zhu, W, Silverman, R, Liu, D. | | Deposit date: | 2022-01-04 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Rational Design, Synthesis, and Mechanism of (3 S ,4 R )-3-Amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylic Acid: Employing a Second-Deprotonation Strategy for Selectivity of Human Ornithine Aminotransferase over GABA Aminotransferase.
J.Am.Chem.Soc., 144, 2022
|
|
8CZU
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 16d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{S})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
6UV7
 
 | |
6AEM
 
 | | Crystal structure of the PKD1 domain of Vibrio anguillarum Epp | | Descriptor: | CALCIUM ION, PKD domain, ZINC ION | | Authors: | Ma, Q, Li, P. | | Deposit date: | 2018-08-05 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.272 Å) | | Cite: | Structural basis for specific calcium binding by the polycystic-kidney-disease domain of Vibrio anguillarum protease Epp
Biochem. Biophys. Res. Commun., 505, 2018
|
|
8PAC
 
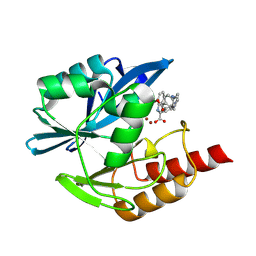 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2553 | | Descriptor: | 7-[(1~{S})-1-[4-(aminomethyl)phenyl]carbonyloxyethyl]-3-[6-(morpholin-4-ylmethyl)pyridin-3-yl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2553
To Be Published
|
|
5BQN
 
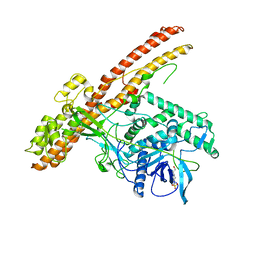 | | Crystal structure of the LHn fragment of botulinum neurotoxin type D, mutant H233Y E230Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Botulinum neurotoxin type D,Botulinum neurotoxin type D | | Authors: | Masuyer, G, Davies, J.R, Moore, K, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of Clostridium botulinum neurotoxin type D as a platform for the development of targeted secretion inhibitors.
Sci Rep, 5, 2015
|
|
8PB0
 
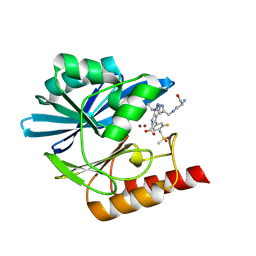 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2696 | | Descriptor: | 7-[(1~{S})-1-[4-[[[(2~{S})-2-azanyl-3-oxidanyl-propanoyl]amino]methyl]-1,2,3-triazol-1-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2696
To Be Published
|
|
7SUY
 
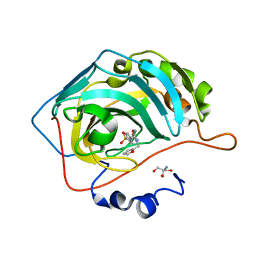 | |
8CZT
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7VU2
 
 | | Chitoporin from Serratia marcescens | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Amornloetwattana, R, Robinson, R.C, van den Berg, B, Suginta, W. | | Deposit date: | 2021-11-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Chitoporin from Serratia marcescens
To Be Published
|
|
8P8F
 
 | |
8CZV
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
6OI2
 
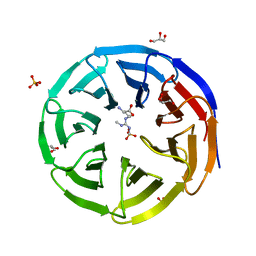 | | Crystal structure of human WDR5 in complex with symmetric dimethyl-L-arginine | | Descriptor: | GLYCEROL, N3, N4-DIMETHYLARGININE, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
5BV7
 
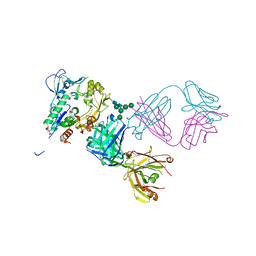 | | Crystal structure of human LCAT (L4F, N5D) in complex with Fab of an agonistic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 27C3 heavy chain, 27C3 light chain, ... | | Authors: | Piper, D.E, Romanow, W.G, Thibault, S.T, Walker, N.P.C. | | Deposit date: | 2015-06-04 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Agonistic Human Antibodies Binding to Lecithin-Cholesterol Acyltransferase Modulate High Density Lipoprotein Metabolism.
J.Biol.Chem., 291, 2016
|
|
6OUJ
 
 | | Carbonic Anhydrase II complexed with benzene sulfonamide MB11-689A | | Descriptor: | 3-[(1S)-1-(4-nitrophenyl)ethyl]-2-oxo-2,3-dihydro-1,3-benzoxazole-5-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Kota, A, McKenna, R. | | Deposit date: | 2019-05-04 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | Carbonic Anhydrase II complexed with benzene sulfonamide MB11-689A
To Be Published
|
|
8PC4
 
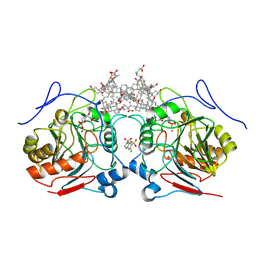 | | MEMBRANE TARGET COMPLEX 1 | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, ... | | Authors: | Garau, G. | | Deposit date: | 2023-06-09 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | MEMBRANE TARGET COMPLEX 1
To Be Published
|
|
6M7L
 
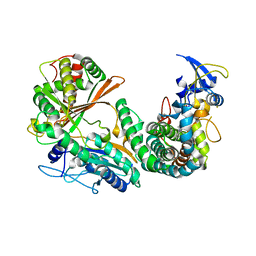 | | Complex of OxyA with the X-domain from GPA biosynthesis | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase, Putative non-ribosomal peptide synthetase | | Authors: | Greule, A, Izore, T, Tailhades, J, Peschke, M, Schoppet, M, Ahmed, I, Kulik, A, Adamek, M, Ziemert, N, De Voss, J, Stegmann, E, Cryle, M.J. | | Deposit date: | 2018-08-20 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.648297 Å) | | Cite: | Kistamicin biosynthesis reveals the biosynthetic requirements for production of highly crosslinked glycopeptide antibiotics.
Nat Commun, 10, 2019
|
|
6UW1
 
 | | The crystal structure of FbiA from Mycobacterium Smegmatis, Fo bound form | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, CALCIUM ION, Phosphoenolpyruvate transferase | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
7SMU
 
 | | Crystal Structure of Consomatin-Ro1 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45-pentadecaoxaoctatetracontane-1,48-diol, Consomatin-Ro1 | | Authors: | Ramiro, I.B.L, Whitby, F.G, Hill, C.P, Safavi-Hemami, H, Concepcion, G.P, Olivera, B.M. | | Deposit date: | 2021-10-26 | | Release date: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Somatostatin venom analogs evolved by fish-hunting cone snails: From prey capture behavior to identifying drug leads.
Sci Adv, 8, 2022
|
|
6VAT
 
 | |
