1E66
 
 | | STRUCTURE OF ACETYLCHOLINESTERASE COMPLEXED WITH (-)-HUPRINE X AT 2.1A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-CHLORO-9-ETHYL-6,7,8,9,10,11-HEXAHYDRO-7,11-METHANOCYCLOOCTA[B]QUINOLIN-12-AMINE, ACETYLCHOLINESTERASE | | Authors: | Dvir, H, Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3D Structure of Torpedo Californica Acetylcholinesterase Complexed with Huprine X at 2. 1 A Resolution: Kinetic and Molecular Dynamic Correlates.
Biochemistry, 41, 2002
|
|
1EMM
 
 | |
1E6K
 
 | | Two-component signal transduction system D12A mutant of CheY | | Descriptor: | Chemotaxis protein CheY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2001-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
1E62
 
 | | Ferredoxin:NADP+ reductase mutant with Lys 75 replaced by Arg (K75R) | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2000-08-07 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners.
Proteins, 59, 2005
|
|
1E9Y
 
 | | Crystal structure of Helicobacter pylori urease in complex with acetohydroxamic acid | | Descriptor: | ACETOHYDROXAMIC ACID, NICKEL (II) ION, UREASE SUBUNIT ALPHA, ... | | Authors: | Ha, N.-C, Oh, S.-T, Oh, B.-H. | | Deposit date: | 2000-11-01 | | Release date: | 2001-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Supramolecular Assembly and Acid Resistance of Helicobacter Pylori Urease
Nat.Struct.Biol., 8, 2001
|
|
1E7O
 
 | | A-SPECTRIN SH3 DOMAIN A11V, V23L, M25V, V44I, V58L MUTATIONS | | Descriptor: | GLYCEROL, SPECTRIN ALPHA CHAIN | | Authors: | Vega, M.C, Serrano, L. | | Deposit date: | 2000-08-31 | | Release date: | 2003-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Thermodynamic and Kinetic Analysis of the Folding Pathway of an SH3 Domain Entropically Stabilised by a Redesigned Hydrophobic Core
J.Mol.Biol., 328, 2003
|
|
1EAW
 
 | |
1E7Y
 
 | | ACTIVE SITE MUTANT (D177->N) OF GLUCOSE 6-PHOSPHATE DEHYDROGENASE FROM LEUCONOSTOC MESENTEROIDES COMPLEXED WITH SUBSTRATE AND NADPH | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CALCIUM ION, GLUCOSE 6-PHOSPHATE 1-DEHYDROGENASE, ... | | Authors: | Adams, M.J, Cosgrove, M.S, Gover, S. | | Deposit date: | 2000-09-12 | | Release date: | 2000-12-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | An Examination of the Role of Asp-177 in the His-Asp Catalytic Dyad of Leuconostoc Mesenteroides Glucose 6-Phosphate Dehydrogenase: X-Ray Structure and Ph Dependence of Kinetic Parameters of the D177N Mutant Enzyme
Biochemistry, 39, 2000
|
|
1E8V
 
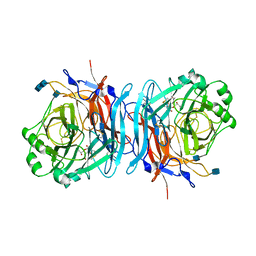 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
1ECL
 
 | |
1ECV
 
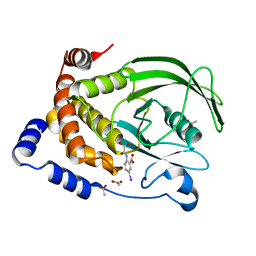 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 5-IODO-2-(OXALYL-AMINO)-BENZOIC ACID | | Descriptor: | 5-IODO-2-(OXALYL-AMINO)-BENZOIC ACID, ACETATE ION, PROTEIN-TYROSINE PHOSPHATASE 1B | | Authors: | Andersen, H.S, Iversen, L.F, Branner, S, Rasmussen, H.B, Moller, N.P.H. | | Deposit date: | 2000-01-26 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 2-(oxalylamino)-benzoic acid is a general, competitive inhibitor of protein-tyrosine phosphatases.
J.Biol.Chem., 275, 2000
|
|
1E9H
 
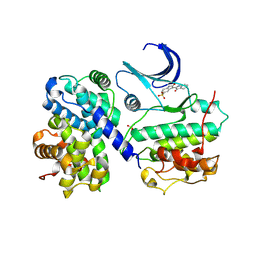 | | Thr 160 phosphorylated CDK2 - Human cyclin A3 complex with the inhibitor indirubin-5-sulphonate bound | | Descriptor: | 2',3-DIOXO-1,1',2',3-TETRAHYDRO-2,3'-BIINDOLE-5'-SULFONIC ACID, CELL DIVISION PROTEIN KINASE 2, CYCLIN A3 | | Authors: | Davies, T.G, Tunnah, P, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2000-10-16 | | Release date: | 2001-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor Binding to Active and Inactive Cdk2: The Crystal Structure of Cdk2-Cyclin A/Indirubin-5-Sulphonate
Structure, 9, 2001
|
|
1ED4
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH IPITU (H4B FREE) | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 2000-01-26 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas.
J.Inorg.Biochem., 81, 2000
|
|
1EAA
 
 | |
1EA4
 
 | | TRANSCRIPTIONAL REPRESSOR COPG/22bp dsDNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*CP*GP*TP*GP *CP*AP*CP*TP*CP*AP*AP*TP*GP*CP*AP*AP*TP*C)-3'), DNA(5'-D(*AP*GP*AP*TP*TP*GP*CP*AP*TP *TP*GP*AP*GP*TP*GP*CP*AP*CP*GP*GP*TP*T)-3'), TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Costa, M, Sola, M, Acebo, P, Eritja, R, Espinosa, M, Solar, G.D, Coll, M. | | Deposit date: | 2000-11-05 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plasmid Transcriptional Repressor Copg Oligomerises to Render Helical Superstructures Unbound and in Complexes with Oligonucleotides
J.Mol.Biol., 310, 2001
|
|
1EFP
 
 | | ELECTRON TRANSFER FLAVOPROTEIN (ETF) FROM PARACOCCUS DENITRIFICANS | | Descriptor: | ADENOSINE MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (ELECTRON TRANSFER FLAVOPROTEIN) | | Authors: | Roberts, D.L, Salazar, D, Fulmer, J.P, Frerman, F.E, Kim, J.J.-P. | | Deposit date: | 1998-12-18 | | Release date: | 1999-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Paracoccus denitrificans electron transfer flavoprotein: structural and electrostatic analysis of a conserved flavin binding domain.
Biochemistry, 38, 1999
|
|
1EKL
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 E35K | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-21 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1EML
 
 | |
1ENQ
 
 | | CO-CRYSTALS OF DEMETALLIZED CONCANAVALIN A WITH ZINC HAVING A ZINC ION BOUND IN THE S1 SITE | | Descriptor: | CONCANAVALIN A, ZINC ION | | Authors: | Bouckaert, J, Loris, R, Poortmans, F, Wyns, L. | | Deposit date: | 1996-03-20 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequential structural changes upon zinc and calcium binding to metal-free concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1EPA
 
 | |
1EPZ
 
 | | CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOASE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH BOUND LIGAND. | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C, Edwards, A.M. | | Deposit date: | 2000-03-30 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
1EQA
 
 | | VACCINIA METHYLTRANSFERASE VP39 MUTANT E233Q COMPLEXED WITH M7G AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 7-METHYLGUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1ETH
 
 | | TRIACYLGLYCEROL LIPASE/COLIPASE COMPLEX | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Hermoso, J, Pignol, D, Kerfelec, B, Crenon, I, Chapus, C, Fontecilla-Camps, J.C. | | Deposit date: | 1995-09-13 | | Release date: | 1996-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex.
J.Biol.Chem., 271, 1996
|
|
1N33
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit bound to codon and near-cognate transfer rna anticodon stem-loop mismatched at the second codon position at the a site with paromomycin | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Murphy IV, F.V, Tarry, M.J, Ramakrishnan, V. | | Deposit date: | 2002-10-25 | | Release date: | 2002-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Selection of tRNA by the Ribosome Requires a Transition from an Open to a Closed Form
Cell(Cambridge,Mass.), 111, 2002
|
|
1ETS
 
 | |
