1V6W
 
 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-4-O-methyl-alpha-D-glucuronosyl-xylobiose | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ENDO-1,4-BETA-D-XYLANASE, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
1TFM
 
 | | CRYSTAL STRUCTURE OF A RIBOSOME INACTIVATING PROTEIN IN ITS NATURALLY INHIBITED FORM | | Descriptor: | 2-AMINO-4-ISOPROPYL-PTERIDINE-6-CARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mishra, V, Bilgrami, S, Paramasivam, M, Yadav, S, Sharma, R.S, Kaur, P, Srinivasan, A, Babu, C.R, Singh, T.P. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE OF A RIBOSOME INACTIVATING PROTEIN IN ITS NATURALLY INHIBITED FORM
To be Published
|
|
1SZ6
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM. CRYSTAL STRUCTURE AT 2.05 A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AZIDE ION, BETA-GALACTOSIDE SPECIFIC LECTIN I B CHAIN, ... | | Authors: | Gabdoulkhakov, A.G, Guhlistova, N.E, Lyashenko, A.V, Krauspenhaar, R, Stoeva, S, Voelter, W, Nikonov, S.V, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2004-04-04 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Viscum album Mistletoe Lectin I in native state at 2.05 A resolution, comparison of structure active site conformation in ricin and in viscumin
TO BE PUBLISHED
|
|
6INU
 
 | |
6INN
 
 | |
6INV
 
 | |
6IWR
 
 | | Crystal structure of GalNAc-T7 with UDP, GalNAc and Mn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MANGANESE (II) ION, N-acetylgalactosaminyltransferase 7, ... | | Authors: | Yu, C, Yin, Y.X. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural basis of carbohydrate transfer activity of UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase 7.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6IWQ
 
 | | Crystal structure of GalNAc-T7 with Mn2+ | | Descriptor: | MANGANESE (II) ION, N-acetylgalactosaminyltransferase 7 | | Authors: | Yu, C, Yin, Y.X. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of carbohydrate transfer activity of UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase 7.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6H0B
 
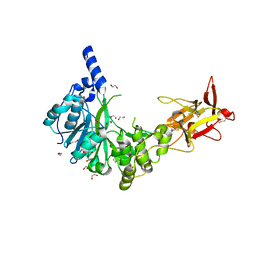 | | Crystal structure of the human GalNAc-T4 in complex with UDP, manganese and the diglycopeptide 6. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, ALA-THR-GLY-ALA-GLY-ALA-GLY-ALA-GLY-THR-THR-PRO-GLY-PRO-GLY, ... | | Authors: | de las Rivas, M, Daniel, E.J.P, Coelho, H, Lira-Navarrete, E, Raich, L, Companon, I, Diniz, A, Lagartera, L, Jimenez-Barbero, J, Clausen, H, Rovira, C, Marcelo, F, Corzana, F, Gerken, T.A, Hurtado-Guerrero, R. | | Deposit date: | 2018-07-08 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Insights into the Catalytic-Domain-Mediated Short-Range Glycosylation Preferences of GalNAc-T4.
ACS Cent Sci, 4, 2018
|
|
6IOE
 
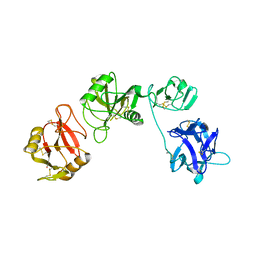 | |
6INO
 
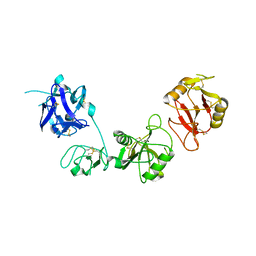 | |
8TFH
 
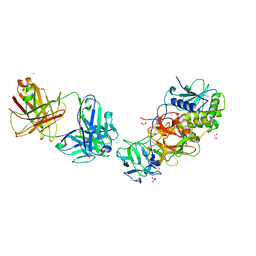 | | Ricin in complex with Fab JB4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, JB4 monoclonal antibody heavy chain, ... | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural Basis of Antibody-Mediated Inhibition of Ricin Toxin Attachment to Host Cells.
Biochemistry, 62, 2023
|
|
8TFL
 
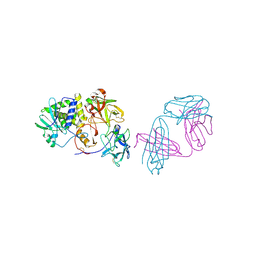 | | Ricin in complex with Fab SylH3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, NONAETHYLENE GLYCOL, ... | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural Basis of Antibody-Mediated Inhibition of Ricin Toxin Attachment to Host Cells.
Biochemistry, 62, 2023
|
|
8V9Q
 
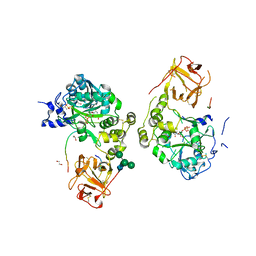 | | Crystal structure of mGalNAc-T1 in complex with the mucin glycopeptide Muc5AC-13, Mn2+, and UDP. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Samara, N.L, Collette, A.M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | An unusual dual sugar-binding lectin domain controls the substrate specificity of a mucin-type O-glycosyltransferase.
Sci Adv, 10, 2024
|
|
8UHV
 
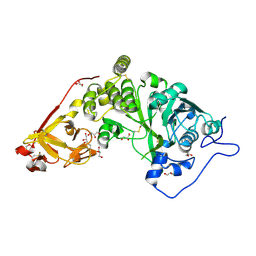 | | X-ray crystal structure of Toxoplasma gondii Apo GalNAc-T3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glycosyl transferase | | Authors: | Kumar, P, Samara, N.L. | | Deposit date: | 2023-10-09 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Toxoplasma gondii O-glycosyltransferase that modulates bradyzoite cyst wall rigidity is distinct from host homologues.
Nat Commun, 15, 2024
|
|
8UJH
 
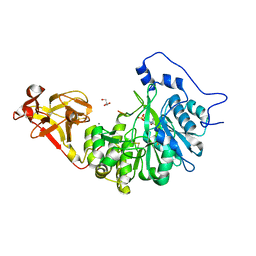 | | X-ray crystal structure of Toxoplasma gondii GalNAc-T3 in complex with UDP-GalNAc, Mn2+, and a CST1 diglycopeptide. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, GLYCEROL, Glycosyl transferase, ... | | Authors: | Kumar, P, Samara, N.L. | | Deposit date: | 2023-10-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Toxoplasma gondii O-glycosyltransferase that modulates bradyzoite cyst wall rigidity is distinct from host homologues.
Nat Commun, 15, 2024
|
|
8UHZ
 
 | |
8UJG
 
 | | X-ray crystal structure of Toxoplasma gondii GalNAc-T3 in complex with UDP-GalNAc, Mn2+, and the mono-glycopeptide Srs13.2 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kumar, P, Samara, N.L. | | Deposit date: | 2023-10-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A Toxoplasma gondii O-glycosyltransferase that modulates bradyzoite cyst wall rigidity is distinct from host homologues.
Nat Commun, 15, 2024
|
|
8UI1
 
 | |
8UJE
 
 | | X-ray crystal structure of Toxoplasma gondii GalNAc-T3 in complex with UDP-GalNAc, Mn2+, and Muc5AC-3,13 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glycosyl transferase, MANGANESE (II) ION, ... | | Authors: | Kumar, P, Samara, N.L. | | Deposit date: | 2023-10-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Toxoplasma gondii O-glycosyltransferase that modulates bradyzoite cyst wall rigidity is distinct from host homologues.
Nat Commun, 15, 2024
|
|
8UJF
 
 | | X-ray crystal structure of Toxoplasma gondii GalNAc-T3 in complex with UDP-GalNAc, Mn2+, and Muc5AC-3,13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyl transferase, MANGANESE (II) ION, ... | | Authors: | Kumar, P, Samara, N.L. | | Deposit date: | 2023-10-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | A Toxoplasma gondii O-glycosyltransferase that modulates bradyzoite cyst wall rigidity is distinct from host homologues.
Nat Commun, 15, 2024
|
|
8UI6
 
 | | X-ray crystal structure of Toxoplasma gondii GalNAc-T3 in complex with UDP-GalNAc, Mn2+, and Muc5AC-3,13 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Glycosyl transferase, MANGANESE (II) ION, ... | | Authors: | Kumar, P, Samara, N.L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Toxoplasma gondii O-glycosyltransferase that modulates bradyzoite cyst wall rigidity is distinct from host homologues.
Nat Commun, 15, 2024
|
|
4ZBV
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with benzyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-15 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
2D23
 
 | | Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | AZIDE ION, ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D7R
 
 | | Crystal structure of pp-GalNAc-T10 complexed with GalNAc-Ser on lectin domain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kubota, T, Shiba, T, Sugioka, S, Kato, R, Wakatsuki, S, Narimatsu, H. | | Deposit date: | 2005-11-25 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10)
J.Mol.Biol., 359, 2006
|
|
