3WD6
 
 | | Crystal structure of Bombyx mori omega-class glutathione transferase in complex with GSH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Yamamoto, K, Suzuki, M, Higashiura, A, Nakagawa, A. | | Deposit date: | 2013-06-07 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of a Bombyx mori Omega-class glutathione transferase.
Biochem.Biophys.Res.Commun., 438, 2013
|
|
3WDR
 
 | | Crystal structure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus complexed with gluco-manno-oligosaccharide | | Descriptor: | BICARBONATE ION, Beta-mannanase, MAGNESIUM ION, ... | | Authors: | Tsukagoshi, H, Ishida, T, Touhara, K.K, Igarashi, K, Samejima, M, Fushinobu, S, Kitamoto, K, Arioka, M. | | Deposit date: | 2013-06-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Family 26 beta-Mannanase from a Symbiotic Protist of the Termite Reticulitermes speratus
J.Biol.Chem., 289, 2014
|
|
1HBH
 
 | |
4CKX
 
 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase N12S mutant (Crystal Form 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
3WPK
 
 | |
3WUA
 
 | |
3WU7
 
 | |
3UO0
 
 | | phosphorylated Bacillus cereus phosphopentomutase soaked with glucose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, ... | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
1M9Z
 
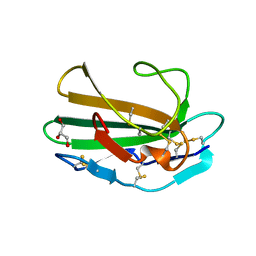 | | CRYSTAL STRUCTURE OF HUMAN TGF-BETA TYPE II RECEPTOR LIGAND BINDING DOMAIN | | Descriptor: | GLYCEROL, TGF-BETA RECEPTOR TYPE II | | Authors: | Boesen, C.C, Radaev, S, Motyka, S.A, Patamawenu, A, Sun, P.D. | | Deposit date: | 2002-07-30 | | Release date: | 2002-09-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | THE 1.1A CRYSTAL STRUCTURE OF HUMAN TGF-BETA TYPE II RECEPTOR LIGAND BINDING DOMAIN
Structure, 10, 2002
|
|
4L3Y
 
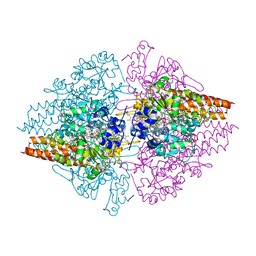 | | Nitrite complex of TvNiR, high dose data set (NO complex) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Lazarenko, V.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2013-06-07 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Investigation of the X-ray-induced nitrite reduction catalysed by cytochrome c nitrite reductase from the bacterium Thioalkalivibrio nitratireducens
To be Published
|
|
3UN5
 
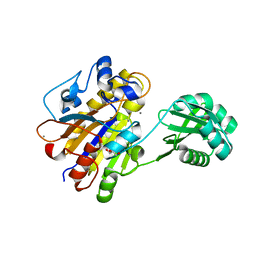 | | Bacillus cereus phosphopentomutase T85E variant | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
4CKY
 
 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | 2,2-dimethyl-3-dehydroquinic acid, 3-DEHYDROQUINATE DEHYDRATASE, SODIUM ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
3UO3
 
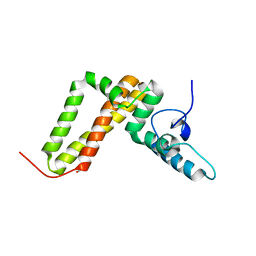 | | Jac1 co-chaperone from Saccharomyces cerevisiae, 5-182 clone | | Descriptor: | ACETATE ION, J-type co-chaperone JAC1, mitochondrial | | Authors: | Osipiuk, J, Bigelow, L, Mulligan, R, Feldmann, B, Babnigg, G, Marszalek, J, Craig, E.A, Dutkiewicz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Interaction of j-protein co-chaperone jac1 with fe-s scaffold isu is indispensable in vivo and conserved in evolution.
J.Mol.Biol., 417, 2012
|
|
3B3C
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine phosphonic acid | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE PHOSPHONIC ACID, POTASSIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-19 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B3S
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE, SODIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3WDQ
 
 | | Crystal structure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-mannanase, MAGNESIUM ION, ... | | Authors: | Tsukagoshi, H, Ishida, T, Touhara, K.K, Igarashi, K, Samejima, M, Fushinobu, S, Kitamoto, K, Arioka, M. | | Deposit date: | 2013-06-20 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Family 26 beta-Mannanase from a Symbiotic Protist of the Termite Reticulitermes speratus
J.Biol.Chem., 289, 2014
|
|
3W2Q
 
 | | EGFR kinase domain T790M/L858R mutant with HKI-272 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, N-(4-{[3-chloro-4-(pyridin-2-ylmethoxy)phenyl]amino}-3-cyano-7-ethoxyquinolin-6-yl)-4-(dimethylamino)butanamide | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
2XJK
 
 | | Monomeric Human Cu,Zn Superoxide dismutase | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE [CU-ZN], ZINC ION | | Authors: | Saraboji, K, Leinartaite, L, Nordlund, A, Oliveberg, M, Logan, D.T. | | Deposit date: | 2010-07-07 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Folding Catalysis by Transient Coordination of Zn2+ to the Cu Ligands of the Als-Associated Enzyme Cu/Zn Superoxide Dismutase 1.
J.Am.Chem.Soc., 132, 2010
|
|
3WAU
 
 | | Crystal structure of 4-O-beta-D-mannosyl-D-glucose phosphorylase MGP complexed with M1P | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-O-phosphono-alpha-D-mannopyranose, 4-O-beta-D-mannosyl-D-glucose phosphorylase, ... | | Authors: | Nakae, S, Ito, S, Higa, M, Senoura, T, Wasaki, J, Hijikata, A, Shionyu, M, Ito, S, Shirai, T. | | Deposit date: | 2013-05-08 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Novel Enzyme in Mannan Biodegradation Process 4-O-beta-d-Mannosyl-d-Glucose Phosphorylase MGP
J.Mol.Biol., 425, 2013
|
|
3UO2
 
 | | Jac1 co-chaperone from Saccharomyces cerevisiae | | Descriptor: | J-type co-chaperone JAC1, mitochondrial | | Authors: | Osipiuk, J, Mulligan, R, Bigelow, L, Marszalek, J, Craig, E.A, Dutkiewicz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Interaction of j-protein co-chaperone jac1 with fe-s scaffold isu is indispensable in vivo and conserved in evolution.
J.Mol.Biol., 417, 2012
|
|
3V0A
 
 | | 2.7 angstrom crystal structure of BoNT/Ai in complex with NTNHA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BoNT/A, CALCIUM ION, ... | | Authors: | Gu, S, Rumpel, S, Zhou, J, Strotmeier, J, Bigalke, H, Perry, K, Shoemaker, C.B, Rummel, A, Jin, R. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Botulinum neurotoxin is shielded by NTNHA in an interlocked complex.
Science, 335, 2012
|
|
5GT8
 
 | | Crystal Structure of apo-CASTOR1 | | Descriptor: | GATS-like protein 3 | | Authors: | Guo, L, Deng, D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of arginine sensor CASTOR1 in arginine-bound and ligand free states
Biochem. Biophys. Res. Commun., 508, 2019
|
|
3V0B
 
 | | 3.9 angstrom crystal structure of BoNT/Ai in complex with NTNHA | | Descriptor: | BoNT/A, CALCIUM ION, NTNH, ... | | Authors: | Gu, S, Rumpel, S, Zhou, J, Strotmeier, J, Bigalke, H, Perry, K, Shoemaker, C.B, Rummel, A, Jin, R. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-14 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Botulinum neurotoxin is shielded by NTNHA in an interlocked complex.
Science, 335, 2012
|
|
3WAS
 
 | | Crystal structure of 4-O-beta-D-mannosyl-D-glucose phosphorylase MGP complexed with Man-Glc+PO4 | | Descriptor: | 4-O-beta-D-mannosyl-D-glucose phosphorylase, PHOSPHATE ION, beta-D-mannopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nakae, S, Ito, S, Higa, M, Senoura, T, Wasaki, J, Hijikata, A, Shionyu, M, Ito, S, Shirai, T. | | Deposit date: | 2013-05-08 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Novel Enzyme in Mannan Biodegradation Process 4-O-beta-d-Mannosyl-d-Glucose Phosphorylase MGP
J.Mol.Biol., 425, 2013
|
|
4CKZ
 
 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
