7NZP
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-fructose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
1CWO
 
 | | HUMAN CYCLOPHILIN A COMPLEXED WITH THR2, LEU5, D-HIV8, LEU10 CYCLOSPORIN | | Descriptor: | CYCLOSPORIN C, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-06-05 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Conformational Differences of an Immunosuppressant Peptolide in a Single Crystal and in a Crystal Complex with Human Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
6X99
 
 | |
1CWI
 
 | | HUMAN CYCLOPHILIN A COMPLEXED WITH 2-VAL 3-(N-METHYL)-D-ALANINE CYCLOSPORIN | | Descriptor: | CYCLOSPORIN D, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-05-25 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structures and Analysis of 11 Cyclosporin Derivatives Complexed with Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
1CWK
 
 | | HUMAN CYCLOPHILIN A COMPLEXED WITH 1-(6,7-DIHYDRO)MEBMT 2-VAL 3-D-(2-S-METHYL)SARCOSINE CYCLOSPORIN | | Descriptor: | CYCLOSPORIN D, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Mikol, V, Kallen, J, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1998-05-26 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures and Analysis of 11 Cyclosporin Derivatives Complexed with Cyclophilin A.
J.Mol.Biol., 283, 1998
|
|
1DPT
 
 | | D-DOPACHROME TAUTOMERASE | | Descriptor: | D-DOPACHROME TAUTOMERASE | | Authors: | Sugimoto, H, Taniguchi, M, Nakagawa, A, Tanaka, I. | | Deposit date: | 1998-05-11 | | Release date: | 1999-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of human D-dopachrome tautomerase, a homologue of macrophage migration inhibitory factor, at 1.54 A resolution.
Biochemistry, 38, 1999
|
|
6PHK
 
 | |
6PHL
 
 | |
7MSE
 
 | | High-resolution crystal structure of hMIF2 with tartrate at the active site | | Descriptor: | D-dopachrome decarboxylase, L(+)-TARTARIC ACID | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
6D4L
 
 | |
7X7W
 
 | | The X-ray Crystallographic Structure of D-Psicose 3-epimerase from Clostridia bacterium | | Descriptor: | D-PSICOSE 3-EPIMERASE, MANGANESE (II) ION | | Authors: | Li, Z.F, Ban, X.F, Xie, X.F, Tian, Y.X, Li, C.M, Gu, Z.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of a novel homodimeric D-allulose 3-epimerase from a Clostridia bacterium
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7C8Q
 
 | | Blasnase-T13A with D-asn | | Descriptor: | Asparaginase, D-ASPARAGINE, FORMIC ACID, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
7SB8
 
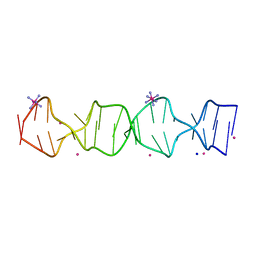 | | d(GA(CGA)5) parallel-stranded homo-duplex | | Descriptor: | COBALT HEXAMMINE(III), GA(CGA)5, SODIUM ION, ... | | Authors: | Luteran, E.M, Paukstelis, P.J. | | Deposit date: | 2021-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.317 Å) | | Cite: | The parallel-stranded d(CGA) duplex is a highly predictable structural motif with two conformationally distinct strands.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6D54
 
 | |
6U1C
 
 | |
1KQ0
 
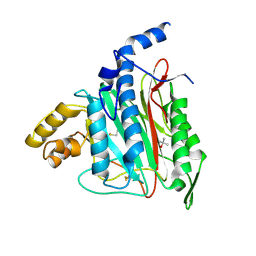 | | Human methionine aminopeptidase type II in complex with D-methionine | | Descriptor: | D-METHIONINE, Methionine aminopeptidase 2, TERTIARY-BUTYL ALCOHOL, ... | | Authors: | Nonato, M.C, Widom, J, Clardy, J. | | Deposit date: | 2002-01-03 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human methionine aminopeptidase type 2 in complex with L- and D-methionine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6N29
 
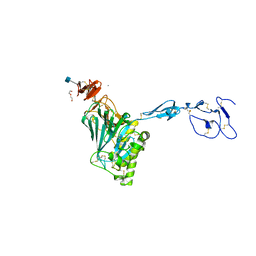 | | Crystal structure of monomeric von Willebrand Factor D`D3 assembly | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dong, X, Arndt, J.W, Springer, T.A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The von Willebrand factor D'D3 assembly and structural principles for factor VIII binding and concatemer biogenesis.
Blood, 133, 2019
|
|
3IJI
 
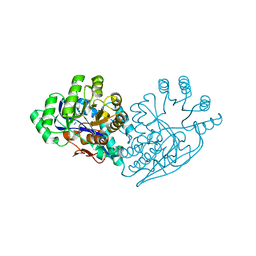 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; nonproductive substrate binding. | | Descriptor: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3IJQ
 
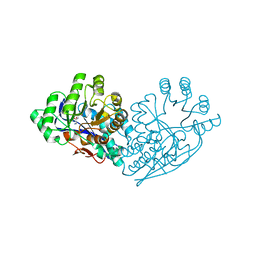 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; productive substrate binding. | | Descriptor: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4QDW
 
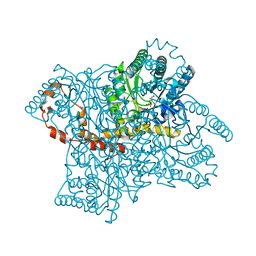 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Ni2+ ions and linear L-arabinose | | Descriptor: | L-arabinose, NICKEL (II) ION, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
4QE5
 
 | |
4QE4
 
 | |
4QEH
 
 | |
4QDP
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Cd2+ ions and cyclic beta-L-arabinose | | Descriptor: | CADMIUM ION, Xylose isomerase, beta-L-arabinopyranose | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
5ZMY
 
 | | Crystal structure of a cis-epoxysuccinate hydrolase producing D(-)-tartaric acids | | Descriptor: | Cis-epoxysuccinate hydrolase, D(-)-TARTARIC ACID, ZINC ION | | Authors: | Dong, S, Liu, X, Wang, X, Feng, Y. | | Deposit date: | 2018-04-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural insight into the catalytic mechanism of a cis-epoxysuccinate hydrolase producing enantiomerically pure d(-)-tartaric acid.
Chem. Commun. (Camb.), 54, 2018
|
|
