3EZY
 
 | | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Dehydrogenase | | Authors: | Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Maletic, M, Gheyi, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-24 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima
To be published
|
|
4K5Z
 
 | | Crystal Structure of Human Chymase in Complex with Fragment Inhibitor 6-chloro-2,3-dihydro-1H-isoindol-1-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-2,3-dihydro-1H-isoindol-1-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
3EXG
 
 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, somatic form, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
2FME
 
 | | Crystal structure of the mitotic kinesin eg5 (ksp) in complex with mg-adp and (r)-4-(3-hydroxyphenyl)-n,n,7,8-tetramethyl-3,4-dihydroisoquinoline-2(1h)-carboxamide | | Descriptor: | (4R)-4-(3-HYDROXYPHENYL)-N,N,7,8-TETRAMETHYL-3,4-DIHYDROISOQUINOLINE-2(1H)-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Sheriff, S. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of human mitotic kinesin Eg5: Characterization of the 4-phenyl-tetrahydroisoquinoline lead series
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FH7
 
 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2I9L
 
 | | Structure of Fab 7D11 from a neutralizing antibody against the poxvirus L1 protein | | Descriptor: | Antibody 7D11 heavy chain, Antibody 7D11 light chain, GLYCEROL, ... | | Authors: | Su, H.P, Golden, J.W, Gittis, A.G, Moss, B, Hooper, J.W, Garboczi, D.N. | | Deposit date: | 2006-09-05 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the binding of the neutralizing antibody, 7D11, to the poxvirus L1 protein
Virology, 368, 2007
|
|
4MKA
 
 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with inhibitor 13 (N-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1,3-benzoxazol-5-yl}methanesulfonamide) | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{3-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1-oxo-1H-isochromen-7-yl}methanesulfonamide, ... | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2013-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a Novel Series of Potent Non-Nucleoside Inhibitors of Hepatitis C Virus NS5B.
J.Med.Chem., 56, 2013
|
|
2POE
 
 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660 | | Descriptor: | Cyclophilin-like protein, putative, FORMIC ACID | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Hassanali, A, Lin, L, Wasney, G, Zhao, Y, Kozieradzki, I, Vedadi, M, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-26 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660.
To be Published
|
|
4K10
 
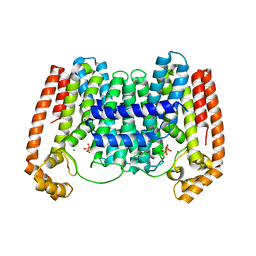 | | Crystal Structure of Leshmaniasis major Farnesyl diphosphate synthase in complex with 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM and Mg2+ | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, ACETATE ION, ... | | Authors: | Aripirala, S, Gabelli, S, Amzel, L.M. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and thermodynamic basis of the inhibition of Leishmania major farnesyl diphosphate synthase by nitrogen-containing bisphosphonates.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2ETL
 
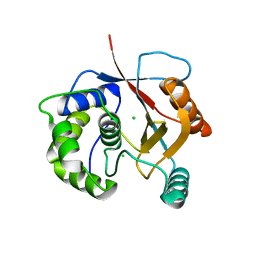 | | Crystal Structure of Ubiquitin Carboxy-terminal Hydrolase L1 (UCH-L1) | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Das, C, Hoang, Q.Q, Kreinbring, C.A, Luchansky, S.J, Meray, R.K, Ray, S.S, Lansbury, P.T, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-10-27 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
4N1D
 
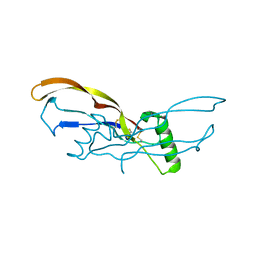 | | Nodal/BMP2 chimera NB250 | | Descriptor: | Nodal/BMP2 chimera protein | | Authors: | Esquivies, L. | | Deposit date: | 2013-10-04 | | Release date: | 2013-12-04 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | Designer Nodal/BMP2 Chimeras Mimic Nodal Signaling, Promote Chondrogenesis, and Reveal a BMP2-like Structure.
J.Biol.Chem., 289, 2014
|
|
3NA1
 
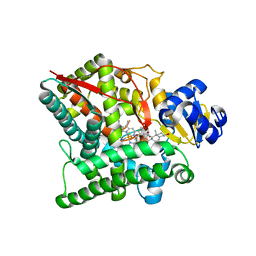 | | Crystal structure of human CYP11A1 in complex with 20-hydroxycholesterol | | Descriptor: | (3alpha,8alpha)-cholest-5-ene-3,20-diol, Adrenodoxin, mitochondrial, ... | | Authors: | Strushkevich, N.V, MacKenzie, F, Tempel, W, Botchkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J.U, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NB0
 
 | |
4N1U
 
 | | Structure of human MTH1 in complex with TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N~4~-cyclopropyl-6-(2,3-dichlorophenyl)pyrimidine-2,4-diamine, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
3EXO
 
 | | Crystal structure of BACE1 bound to inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{2-methyl-5-[(6-phenylpyrimidin-4-yl)amino]phenyl}methanesulfonamide, ... | | Authors: | Allison, T.J. | | Deposit date: | 2008-10-16 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a small molecule beta-secretase inhibitor that binds without catalytic aspartate engagement.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4K3O
 
 | | E. coli sliding clamp in complex with AcQADLF | | Descriptor: | (ACE)QADLF, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
4KLD
 
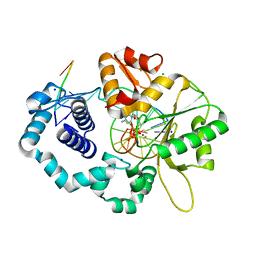 | | DNA polymerase beta matched substrate complex with Ca2+, 0 s | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Freudenthal, B.D, Beard, W.A, Shock, D.D, Wilson, S.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.916 Å) | | Cite: | Observing a DNA polymerase choose right from wrong.
Cell(Cambridge,Mass.), 154, 2013
|
|
4O9Y
 
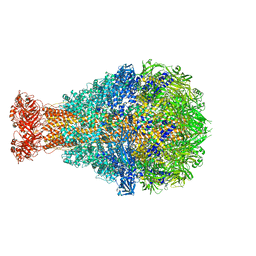 | | Crystal Structure of TcdA1 | | Descriptor: | TcdA1 | | Authors: | Meusch, D, Gatsogiannis, C, Efremov, R.G, Lang, A.E, Hofnagel, O, Vetter, I.R, Aktories, K, Raunser, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Mechanism of Tc toxin action revealed in molecular detail.
Nature, 508, 2014
|
|
4OKS
 
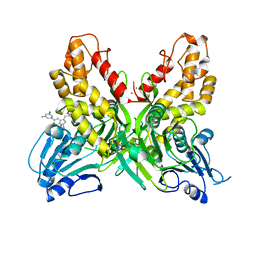 | |
4KM6
 
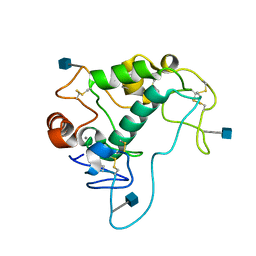 | | Human folate receptor alpha (FOLR1) at acidic pH, orthorhombic form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Folate receptor alpha | | Authors: | Singh, M, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2HYO
 
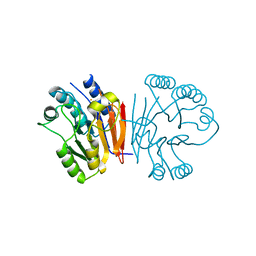 | | Crystal structure of Rv0805 N97A mutant | | Descriptor: | FE (III) ION, MANGANESE (II) ION, Rv0805N97A | | Authors: | Shenoy, A.R, Capuder, M, Draskovic, P, Lamba, D, Visweswariah, S.S, Podobnik, M. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and Biochemical Analysis of the Rv0805 Cyclic Nucleotide Phosphodiesterase from Mycobacterium tuberculosis.
J.Mol.Biol., 365, 2007
|
|
3EYU
 
 | |
4K6K
 
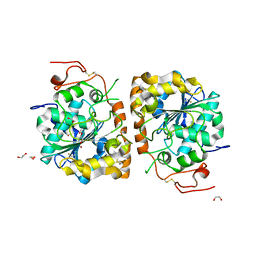 | | Crystal structure of CALB mutant D223G from Candida antarctica | | Descriptor: | 1,2-ETHANEDIOL, Lipase B | | Authors: | An, J, Xie, Y, Feng, Y, Wu, G. | | Deposit date: | 2013-04-16 | | Release date: | 2014-01-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced enzyme kinetic stability by increasing rigidity within the active site.
J.Biol.Chem., 289, 2014
|
|
3NKN
 
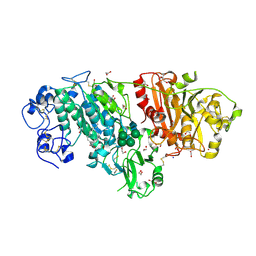 | | Crystal structure of mouse autotaxin in complex with 14:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
2EXR
 
 | | X-Ray Structure of Cytokinin Oxidase/Dehydrogenase (CKX) From Arabidopsis Thaliana AT5G21482 | | Descriptor: | Cytokinin dehydrogenase 7, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-11-08 | | Release date: | 2005-11-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal structure of Arabidopsis thaliana cytokinin dehydrogenase.
Proteins, 70, 2008
|
|
