3BLO
 
 | | TGT mutant in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-12-11 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
3BLD
 
 | | tRNA guanine transglycosylase V233G mutant preQ1 complex structure | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-12-11 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
3BLL
 
 | | TGT mutant in complex with Boc-preQ1 | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION, ... | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-12-11 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
2Z1V
 
 | | tRNA guanine transglycosylase E235Q mutant apo structure, pH 8.5 | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Tidten, N, Stengl, B, Heine, A, Garcia, G.A, Klebe, G, Reuter, K. | | Deposit date: | 2007-05-15 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Glutamate versus Glutamine Exchange Swaps Substrate Selectivity in tRNA-Guanine Transglycosylase: Insight into the Regulation of Substrate Selectivity by Kinetic and Crystallographic Studies
J.Mol.Biol., 374, 2007
|
|
3BL3
 
 | | tRNA guanine transglycosylase V233G mutant apo structure | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-12-10 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
2Z1W
 
 | | tRNA guanine transglycosylase TGT E235Q mutant in complex with BDI (2-BUTYL-5,6-DIHYDRO-1H-IMIDAZO[4,5-D]PYRIDAZINE-4,7-DIONE) | | Descriptor: | 2-BUTYL-5,6-DIHYDRO-1H-IMIDAZO[4,5-D]PYRIDAZINE-4,7-DIONE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Stengl, B, Heine, A, Garcia, G.A, Klebe, G, Reuter, K. | | Deposit date: | 2007-05-16 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Glutamate versus Glutamine Exchange Swaps Substrate Selectivity in tRNA-Guanine Transglycosylase: Insight into the Regulation of Substrate Selectivity by Kinetic and Crystallographic Studies
J.Mol.Biol., 374, 2007
|
|
1T0M
 
 | | Conformational switch in polymorphic H-2K molecules containing an HSV peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Webb, A.I, Borg, N.A, Dunstone, M.A, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Carbone, F.R, Bottomley, S.P, Purcell, A.W, Rossjohn, J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of H-2K(b) and K(bm8) complexed to a herpes simplex virus determinant: evidence for a conformational switch that governs T cell repertoire selection and viral resistance.
J Immunol., 173, 2004
|
|
1T0N
 
 | | Conformational switch in polymorphic H-2K molecules containing an HSV peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Webb, A.I, Borg, N.A, Dunstone, M.A, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Carbone, F.R, Bottomley, S.P, Purcell, A.W, Rossjohn, J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of H-2K(b) and K(bm8) complexed to a herpes simplex virus determinant: evidence for a conformational switch that governs T cell repertoire selection and viral resistance.
J Immunol., 173, 2004
|
|
1KXA
 
 | |
1K9B
 
 | | Crystal structure of the bifunctional soybean Bowman-Birk inhibitor at 0.28 nm resolution. Structural peculiarities in a folded protein conformation | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR | | Authors: | Voss, R.H, Ermler, U, Essen, L.O, Wenzl, G, Kim, Y.M, Flecker, P. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the bifunctional soybean Bowman-Birk inhibitor at 0.28-nm resolution. Structural peculiarities in a folded protein conformation.
Eur.J.Biochem., 242, 1996
|
|
1D6R
 
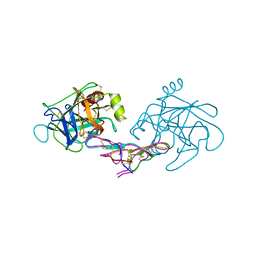 | | CRYSTAL STRUCTURE OF CANCER CHEMOPREVENTIVE BOWMAN-BIRK INHIBITOR IN TERNARY COMPLEX WITH BOVINE TRYPSIN AT 2.3 A RESOLUTION. STRUCTURAL BASIS OF JANUS-FACED SERINE PROTEASE INHIBITOR SPECIFICITY | | Descriptor: | BOWMAN-BIRK PROTEINASE INHIBITOR PRECURSOR, TRYPSINOGEN | | Authors: | Koepke, J, Ermler, U, Wenzl, G, Flecker, P. | | Deposit date: | 1999-10-15 | | Release date: | 2000-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of cancer chemopreventive Bowman-Birk inhibitor in ternary complex with bovine trypsin at 2.3 A resolution. Structural basis of Janus-faced serine protease inhibitor specificity.
J.Mol.Biol., 298, 2000
|
|
8ZUB
 
 | | The Crystal structure of mol075 bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-pentyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]pyrimidine-2,4-dione | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2024-06-08 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of pyrimidone derivatives as nonpeptidic and noncovalent 3-chymotrypsin-like protease (3CL pro ) inhibitors with anti-coronavirus activities.
Bioorg.Chem., 154, 2025
|
|
8ZUC
 
 | | The Crystal structure of mol080 bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[[6-chloranyl-2-(3-methylbutyl)indazol-5-yl]amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]pyrimidine-2,4-dione | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2024-06-08 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of pyrimidone derivatives as nonpeptidic and noncovalent 3-chymotrypsin-like protease (3CL pro ) inhibitors with anti-coronavirus activities.
Bioorg.Chem., 154, 2025
|
|
8ZT9
 
 | | The Crystal structure of mol066 bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-propan-2-yl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]pyrimidine-2,4-dione, GLYCEROL | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2024-06-06 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of pyrimidone derivatives as nonpeptidic and noncovalent 3-chymotrypsin-like protease (3CL pro ) inhibitors with anti-coronavirus activities.
Bioorg.Chem., 154, 2025
|
|
1KXC
 
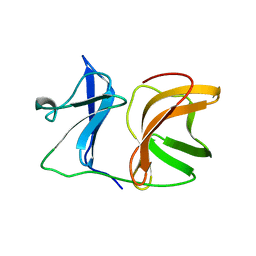 | |
1KXD
 
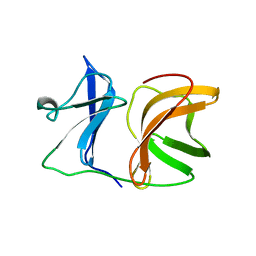 | |
1KXB
 
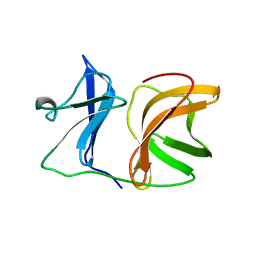 | |
1KXE
 
 | |
1KXF
 
 | |
4LXD
 
 | | Bcl_2-Navitoclax Analog (without Thiophenyl) Complex | | Descriptor: | 4-(4-{[4-(4-chlorophenyl)-5,6-dihydro-2H-pyran-3-yl]methyl}piperazin-1-yl)-N-{[3-nitro-4-(tetrahydro-2H-pyran-4-ylamino)phenyl]sulfonyl}benzamide, Apoptosis regulator Bcl-2 | | Authors: | Park, C.H. | | Deposit date: | 2013-07-29 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets.
NAT.MED. (N.Y.), 19, 2013
|
|
1HAV
 
 | | HEPATITIS A VIRUS 3C PROTEINASE | | Descriptor: | CHLORIDE ION, HEPATITIS A VIRUS 3C PROTEINASE | | Authors: | Bergmann, E.M, James, M.N.G. | | Deposit date: | 1996-10-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition.
J.Virol., 71, 1997
|
|
1KZX
 
 | | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) | | Descriptor: | INTESTINAL FATTY ACID-BINDING PROTEIN (T54) | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) that is associated with altered lipid metabolism
Biochemistry, 42, 2003
|
|
3FTR
 
 | |
5JOD
 
 | | Structure of proplasmepsin IV from Plasmodium falciparum | | Descriptor: | GLYCEROL, Proplasmepsin IV | | Authors: | Recacha, R, Akopjana, I, Tars, K, Jaudzems, K. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Crystal structure of Plasmodium falciparum proplasmepsin IV: the plasticity of proplasmepsins.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7F29
 
 | |
