6LQV
 
 | |
6LR2
 
 | |
6LR7
 
 | |
6LSR
 
 | | Cryo-EM structure of a pre-60S ribosomal subunit - state B | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-20 | | Release date: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
6LU8
 
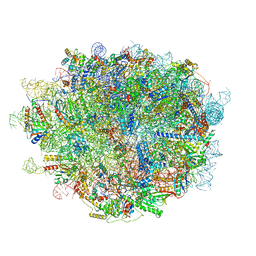 | | Cryo-EM structure of a human pre-60S ribosomal subunit - state A | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-26 | | Release date: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
6LZ2
 
 | | Crystal structure of a thermostable green fluorescent protein (TGP) with a synthetic nanobody (Sb44) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Cai, H, Yao, H, Li, T, Hutter, C, Tang, Y, Li, Y, Seeger, M, Li, D. | | Deposit date: | 2020-02-17 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | An improved fluorescent tag and its nanobodies for membrane protein expression, stability assay, and purification.
Commun Biol, 3, 2020
|
|
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | Authors: | Zhou, Z, Chen, S, Wang, L, Chu, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
6M9X
 
 | |
6M9Y
 
 | |
6M9Z
 
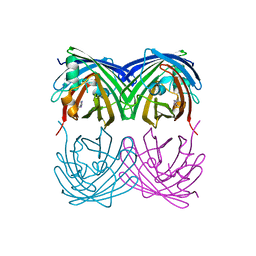 | |
6MAS
 
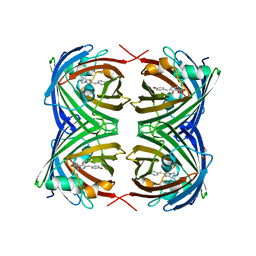 | | X-ray Structure of Branchiostoma floridae fluorescent protein lanFP10G | | Descriptor: | GLYCEROL, Uncharacterized protein | | Authors: | Muslinkina, L, Pletneva, N, Pletnev, V, Pletnev, S. | | Deposit date: | 2018-08-28 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Factors Enabling Successful GFP-Like Proteins with Alanine as the Third Chromophore-Forming Residue.
J. Mol. Biol., 431, 2019
|
|
6MB2
 
 | | Cryo-EM structure of the PYD filament of AIM2 | | Descriptor: | Green fluorescent protein, Interferon-inducible protein AIM2 | | Authors: | Lu, A, Li, Y, Wu, H. | | Deposit date: | 2018-08-29 | | Release date: | 2018-09-05 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2.
Cell Discov, 1, 2015
|
|
6MDR
 
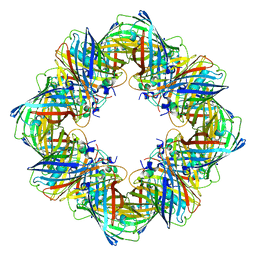 | | Cryo-EM structure of the Ceru+32/GFP-17 protomer | | Descriptor: | Ceru+32, GFP-17 | | Authors: | Simon, A.J, Zhou, Y, Ramasubramani, V, Glaser, J, Pothukuchy, A, Golihar, J, Gerberich, J.C, Leggere, J.C, Morrow, B.R, Jung, C, Glotzer, S.C, Taylor, D.W, Ellington, A.D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Supercharging enables organized assembly of synthetic biomolecules.
Nat Chem, 11, 2019
|
|
6MGH
 
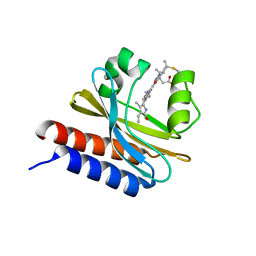 | | X-ray structure of monomeric near-infrared fluorescent protein miRFP670nano | | Descriptor: | 3-[2-[(~{Z})-[5-[(~{Z})-[(3~{S},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Pletnev, S. | | Deposit date: | 2018-09-13 | | Release date: | 2018-12-19 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing.
Nat Commun, 10, 2019
|
|
6MHT
 
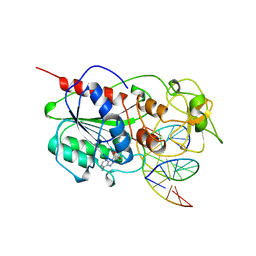 | |
6MKS
 
 | | Cryo-EM structure of NLRC4-CARD filament | | Descriptor: | Chimera protein of NLR family CARD domain-containing protein 4 and EGFP | | Authors: | Zheng, W, Matyszewski, M, Sohn, J, Egelman, E.H. | | Deposit date: | 2018-09-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the NLRC4CARDfilament provides insights into how symmetric and asymmetric supramolecular structures drive inflammasome assembly.
J. Biol. Chem., 293, 2018
|
|
6MLT
 
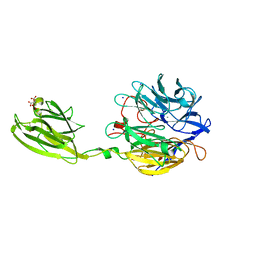 | | Crystal structure of the V. cholerae biofilm matrix protein Bap1 | | Descriptor: | CALCIUM ION, CITRATE ANION, GLYCEROL, ... | | Authors: | Kaus, K, Biester, A, Chupp, E, Lu, K, Vidsudharomn, C, Olson, R. | | Deposit date: | 2018-09-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 angstrom crystal structure of the extracellular matrix protein Bap1 fromVibrio choleraeprovides insights into bacterial biofilm adhesion.
J.Biol.Chem., 294, 2019
|
|
6MRN
 
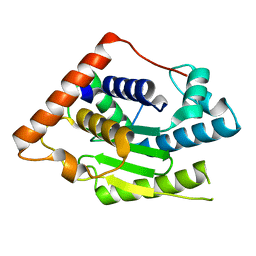 | | Crystal Structure of ChlaDUB2 DUB domain | | Descriptor: | Deubiquitinase and deneddylase Dub2 | | Authors: | Hausman, J.M, Das, C. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Two Deubiquitinating Enzymes fromChlamydia trachomatisHave Distinct Ubiquitin Recognition Properties.
Biochemistry, 59, 2020
|
|
6MTB
 
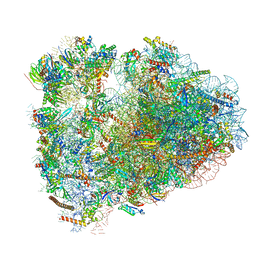 | | Rabbit 80S ribosome with P- and Z-site tRNAs (unrotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6MTC
 
 | | Rabbit 80S ribosome with Z-site tRNA and IFRD2 (unrotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6MTD
 
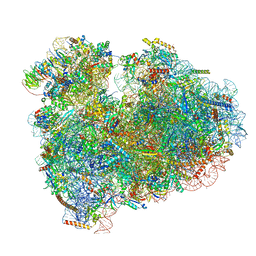 | | Rabbit 80S ribosome with eEF2 and SERBP1 (unrotated state with 40S head swivel) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6MTE
 
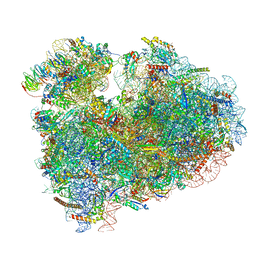 | | Rabbit 80S ribosome with eEF2 and SERBP1 (rotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6MWQ
 
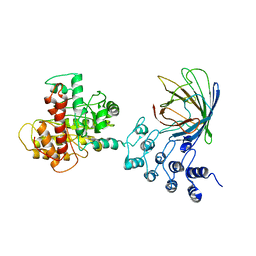 | |
6MZ3
 
 | | mCherry pH sensitive mutant - M66T (mCherryTYG) | | Descriptor: | PAmCherry1 protein | | Authors: | Haynes, E.P, Tantama, M. | | Deposit date: | 2018-11-03 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.088 Å) | | Cite: | Quantifying Acute Fuel and Respiration Dependent pH Homeostasis in Live Cells Using the mCherryTYG Mutant as a Fluorescence Lifetime Sensor.
Anal.Chem., 91, 2019
|
|
6NHT
 
 | |
