4AZA
 
 | | Improved eIF4E binding peptides by phage display guided design. | | Descriptor: | EIF4G1_D5S PEPTIDE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, [[(2R,3S,4R,5R)-5-(6-AMINO-3-METHYL-4-OXO-5H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHOXY-HYDROXY-PHOSPHORYL] PHOSPHONO HYDROGEN PHOSPHATE | | Authors: | Chew, W.Z, Quah, S.T, Verma, C.S, Liu, Y, Lane, D.P, Brown, C.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Improved Eif4E Binding Peptides by Phage Display Guided Design: Plasticity of Interacting Surfaces Yield Collective Effects.
Plos One, 7, 2012
|
|
4B88
 
 | |
3MC0
 
 | | Crystal Structure of Staphylococcal Enterotoxin G (SEG) in Complex with a Mouse T-cell Receptor beta Chain | | Descriptor: | ACETATE ION, Enterotoxin SEG, variable beta 8.2 mouse T cell receptor | | Authors: | Fernandez, M.M, Cho, S, Robinson, H, Mariuzza, R.A, Malchiodi, E.L. | | Deposit date: | 2010-03-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of staphylococcal enterotoxin G (SEG) in complex with a mouse T-cell receptor {beta} chain.
J.Biol.Chem., 286, 2011
|
|
3MCG
 
 | |
3MA3
 
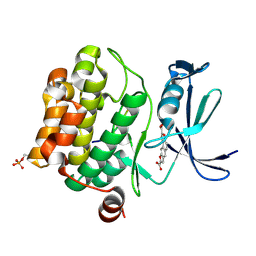 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a naphtho-difuran ligand | | Descriptor: | Pimtide, Proto-oncogene serine/threonine-protein kinase pim-1, naphtho[2,1-b:7,6-b']difuran-2,8-dicarboxylic acid | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Vollmar, M, von Delft, F, Cochet, C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New potent dual inhibitors of CK2 and Pim kinases: discovery and structural insights.
Faseb J., 24, 2010
|
|
4BI1
 
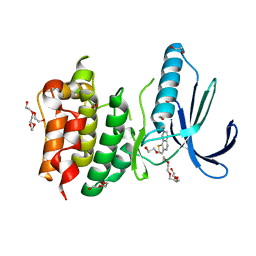 | | Scaffold Focused Virtual Screening: Prospective Application to the Discovery of TTK Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Langdon, S.R, Westwood, I.M, van Montfort, R.L.M, Brown, N, Blagg, J. | | Deposit date: | 2013-04-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Scaffold-Focused Virtual Screening: Prospective Application to the Discovery of Ttk Inhibitors.
J.Chem.Inf.Model, 53, 2013
|
|
4BLA
 
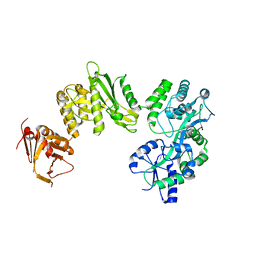 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form II) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3MC9
 
 | |
7P72
 
 | | The PDZ domain of SNX27 complexed with the PDZ-binding motif of MERS-E | | Descriptor: | CALCIUM ION, Envelope small membrane protein, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
4AXS
 
 | | Structure of Carbamate Kinase from Mycoplasma penetrans | | Descriptor: | CARBAMATE KINASE, SULFATE ION | | Authors: | Gallego, P, Planell, R, Benach, J, Querol, E, PerezPons, J.A, Reverter, D. | | Deposit date: | 2012-06-14 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma Penetrans.
Plos One, 7, 2012
|
|
4AY7
 
 | | methyltransferase from Methanosarcina mazei | | Descriptor: | MAGNESIUM ION, METHYLCOBALAMIN: COENZYME M METHYLTRANSFERASE, ZINC ION | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeld, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5AVT
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
7P9W
 
 | | Epstein-Barr virus encoded apoptosis regulator BHRF1 in complex with Puma BH3 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, AMMONIUM ION, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.00010061 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
5AW3
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 100 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
3N8V
 
 | | Crystal Structure of Unoccupied Cyclooxygenase-1 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Sidhu, R.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
4BAC
 
 | | prototype foamy virus strand transfer complexes on product DNA | | Descriptor: | 5'-D(*AP*GP*GP*AP*GP*CP*CP*AP*AP*GP*AP*CP*GP*GP *AP*TP*CP)-3', 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*TP*GP*TP*AP)-3', DNA (38-MER), ... | | Authors: | Yin, Z, Lapkouski, M, Yang, W, Craigie, R. | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | Assembly of Prototype Foamy Virus Strand Transfer Complexes on Product DNA Bypassing Catalysis of Integration.
Protein Sci., 21, 2012
|
|
3MLY
 
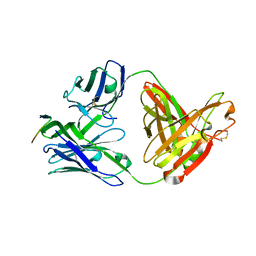 | | Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with a UR29 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4BHD
 
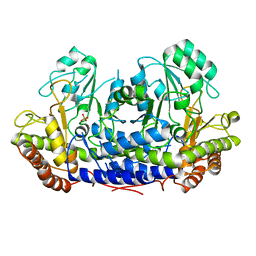 | |
3MLV
 
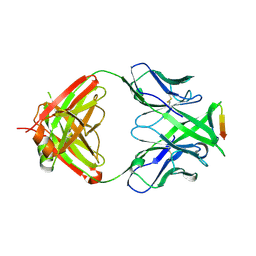 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with an NOF V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7P4O
 
 | | Crystal structure of Autotaxin and 9(R)-delta6a,10a-THC | | Descriptor: | (9~{R},10~{a}~{S})-6,6,9-trimethyl-3-pentyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A, Hausmann, J. | | Deposit date: | 2021-07-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Linking medicinal cannabis to autotaxin-lysophosphatidic acid signaling.
Life Sci Alliance, 6, 2023
|
|
7P4J
 
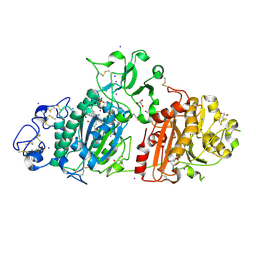 | | Crystal structure of Autotaxin and tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A, Hausmann, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Linking medicinal cannabis to autotaxin-lysophosphatidic acid signaling.
Life Sci Alliance, 6, 2023
|
|
7OYP
 
 | | Carbonic anhydrase II in complex with Hit3-t1 (MH172) | | Descriptor: | (2S)-3-oxidanyl-2-[2-[(4-sulfamoylphenyl)methoxyamino]ethanoylamino]propanamide, 4-methylbenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2021-06-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Identification of specific carbonic anhydrase inhibitors via in situ click chemistry, phage-display and synthetic peptide libraries: comparison of the methods and structural study.
Rsc Med Chem, 14, 2023
|
|
3MQG
 
 | | crystal structure of the 3-N-acetyl transferase WlbB from Bordetella petrii in complex with acetyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETYL COENZYME *A, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Molecular structure of WlbB, a bacterial N-acetyltransferase involved in the biosynthesis of 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid .
Biochemistry, 49, 2010
|
|
7OYN
 
 | | Carbonic anhydrase II in complex with Hit3 (MH57) | | Descriptor: | Carbonic anhydrase 2, Hit3 (MH57), ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2021-06-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Identification of specific carbonic anhydrase inhibitors via in situ click chemistry, phage-display and synthetic peptide libraries: comparison of the methods and structural study.
Rsc Med Chem, 14, 2023
|
|
3MRQ
 
 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with Melan-A MART1 decapeptide variant | | Descriptor: | 10-meric peptide from Melanoma antigen recognized by T-cells 1, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Reiser, J.-B, Le Gorrec, M, Chouquet, A, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of MHC class I HLA-A2 molecule complexed with Melan-A MART1 decapeptide variant
To be Published
|
|
