5LA5
 
 | |
8G4Y
 
 | | Structure of ZNRF3 ECD bound to peptide MK1-3.6.10 | | Descriptor: | E3 ubiquitin-protein ligase ZNRF3, MK1-3.6.10 | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2023-02-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Potent and selective binders of the E3 ubiquitin ligase ZNRF3 stimulate Wnt signaling and intestinal organoid growth.
Cell Chem Biol, 31, 2024
|
|
6VTJ
 
 | |
5L7G
 
 | | MCR IN COMPLEX WITH ligand | | Descriptor: | 1,2-ETHANEDIOL, Mineralocorticoid receptor, NCOA1 peptide, ... | | Authors: | Edman, K, Aagaard, A, Backstrom, S, Xue, Y. | | Deposit date: | 2016-06-03 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Drug Design of Mineralocorticoid Receptor Antagonists to Explore Oxosteroid Receptor Selectivity.
ChemMedChem, 12, 2017
|
|
5L7N
 
 | | Plexin A1 extracellular fragment, domains 7-10 (IPT3-IPT6) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kong, Y, Janssen, B.J.C, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
8GLZ
 
 | | Crystal structure of T252E-CYP199A4 in complex with 4-hydroxybenzoic acid. Crystal was initially co-crystallised with 4-methoxybenzoic acid and soaked with 4 mM hydrogen peroxide | | Descriptor: | CHLORIDE ION, Cytochrome P450, P-HYDROXYBENZOIC ACID, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An In Crystallo Reaction with an Engineered Cytochrome P450 Peroxygenase.
Chemistry, 30, 2024
|
|
5LAU
 
 | | Oceanobacillus iheyensis macrodomain mutant G37V with ADPR | | Descriptor: | GLYCEROL, MacroD-type macrodomain, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
5L7X
 
 | | Afamin antibody fragment, N14 Fab, L1- glycosylated, crystal form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Mouse Antibody Fab Fragment, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-06-04 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4YHE
 
 | | NATIVE BACTEROIDETES-AFFILIATED GH5 CELLULASE LINKED WITH A POLYSACCHARIDE UTILIZATION LOCUS | | Descriptor: | GH5 | | Authors: | Naas, A.E, MacKenzie, A.K, Dalhus, B, Eijsink, V.G.H, Pope, P.B. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Features of a Bacteroidetes-Affiliated Cellulase Linked with a Polysaccharide Utilization Locus.
Sci Rep, 5, 2015
|
|
5L89
 
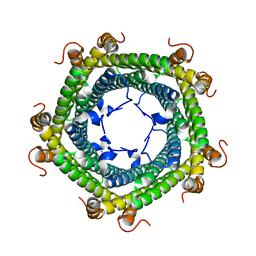 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A | | Descriptor: | CALCIUM ION, Rru_A0973 | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
8FUL
 
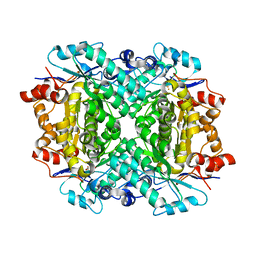 | |
8GM2
 
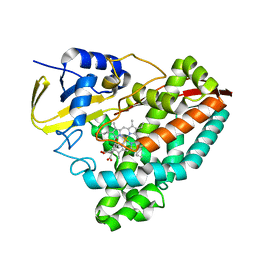 | |
8FK8
 
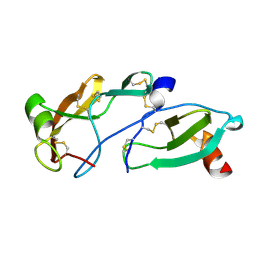 | | Crystal Structure of the Tick Evasin EVA-AAM1001(L39P) Complexed to Human Chemokine CCL7 | | Descriptor: | C-C motif chemokine 7, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
5KPU
 
 | |
8GBE
 
 | |
6VW8
 
 | | Formate Dehydrogenase FdsABG subcomplex FdsBG from C. necator | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Young, T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and kinetic analyses of the FdsBG subcomplex of the cytosolic formate dehydrogenase FdsABG fromCupriavidus necator.
J.Biol.Chem., 295, 2020
|
|
5KQW
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobutyrate transaminase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
8FK6
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001(Y44A) Complexed to Human Chemokine CCL7 | | Descriptor: | C-C motif chemokine 7, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
5L9Y
 
 | | Crystal structure of human heparanase, in complex with glucuronic acid configured aziridine probe JJB355 | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{S},6~{R})-2-(8-azidooctylamino)-3,4,5,6-tetrakis(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
8GOD
 
 | | Co-crystal structure of Human Protein-arginine deiminase type-4 (PAD4) with small molecule inhibitor JBI-589 | | Descriptor: | Protein-arginine deiminase type-4, [(3~{R})-3-azanylpiperidin-1-yl]-[2-[1-[(4-fluorophenyl)methyl]indol-2-yl]-3-methyl-imidazo[1,2-a]pyridin-7-yl]methanone | | Authors: | Swaminathan, S, Birudukota, S, Vaithilingam, K, Kandan, S, Asaithambi, K, Kathiresan, N, Gosu, R, Rajagopal, S, Sadhu, N. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Alleviation of arthritis through prevention of neutrophil extracellular traps by an orally available inhibitor of protein arginine deiminase 4.
Sci Rep, 13, 2023
|
|
8G6P
 
 | | Crystal structure of Mycobacterium thermoresistibile MurE in complex with ADP and 2,6-Diaminopimelic acid | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Rossini, N.O, Silva, C.S, Dias, M.V.B. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of Mycobacterium thermoresistibile MurE ligase reveals the binding mode of the substrate m-diaminopimelate.
J.Struct.Biol., 215, 2023
|
|
8FH4
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undec-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile | | Descriptor: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-13 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FKO
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(2S,5R)-5-Amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | Descriptor: | (3P)-3-{4-[(2S,5R)-5-amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-21 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8GAI
 
 | |
5KSP
 
 | | hMiro1 C-domain GDP Complex C2221 Crystal Form | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
