5E4M
 
 | |
5WFK
 
 | |
5WF0
 
 | |
5EBH
 
 | |
4ME6
 
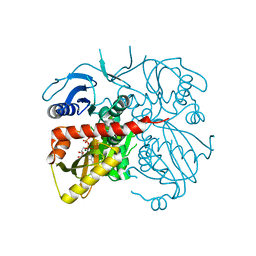 | | Crystal structure of D-alanine-D-alanine ligase A from Xanthomonas oryzae pathovar oryzae with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine--D-alanine ligase, MAGNESIUM ION | | Authors: | Doan, T.T.N, Kim, J.K, Ahn, Y.J, Lee, B.M, Kang, L.W. | | Deposit date: | 2013-08-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of d-alanine-d-alanine ligase from Xanthomonas oryzae pv. oryzae alone and in complex with nucleotides
Arch.Biochem.Biophys., 545C, 2014
|
|
7K07
 
 | |
6U6P
 
 | |
6U6R
 
 | |
6U6S
 
 | |
6OO7
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in nanodiscs | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
5LD2
 
 | | Cryo-EM structure of RecBCD+DNA complex revealing activated nuclease domain | | Descriptor: | Fork-Hairpin DNA (70-MER), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wilkinson, M, Chaban, Y, Wigley, D.B. | | Deposit date: | 2016-06-23 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Mechanism for nuclease regulation in RecBCD.
Elife, 5, 2016
|
|
1M2Z
 
 | | Crystal structure of a dimer complex of the human glucocorticoid receptor ligand-binding domain bound to dexamethasone and a TIF2 coactivator motif | | Descriptor: | DEXAMETHASONE, glucocorticoid receptor, nuclear receptor coactivator 2, ... | | Authors: | Bledsoe, R.B, Montana, V.G, Stanley, T.B, Delves, C.J, Apolito, C.J, Mckee, D.D, Consler, T.G, Parks, D.J, Stewart, E.L, Willson, T.M, Lambert, M.H, Moore, J.T, Pearce, K.H, Xu, H.E. | | Deposit date: | 2002-06-26 | | Release date: | 2003-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Glucocorticoid Receptor Ligand Binding Domain Reveals a Novel Mode of Receptor Dimerization and Coactivator Recognition
Cell(Cambridge,Mass.), 110, 2002
|
|
5LFH
 
 | | NMR structure of peptide 10 targeting CXCR4 | | Descriptor: | ACE-ARG-ALA-DCY-ARG-PHE-PHE-CYS | | Authors: | Di Maro, S, Trotta, A.M, Brancaccio, D, Di Leva, F.S, La Pietra, V, Ierano, C, Napolitano, M, Portella, L, D'Alterio, C, Siciliano, R.A, Sementa, D, Tomassi, S, Carotenuto, A, Novellino, E, Scala, S, Marinelli, L. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2016-10-05 | | Method: | SOLUTION NMR | | Cite: | Exploring the N-Terminal Region of C-X-C Motif Chemokine 12 (CXCL12): Identification of Plasma-Stable Cyclic Peptides As Novel, Potent C-X-C Chemokine Receptor Type 4 (CXCR4) Antagonists.
J.Med.Chem., 59, 2016
|
|
5LFN
 
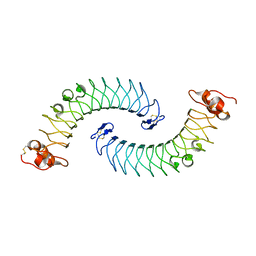 | | Crystal structure of human chondroadherin | | Descriptor: | CHLORIDE ION, Chondroadherin | | Authors: | Ramisch, S, Pramhed, A, Logan, D.T. | | Deposit date: | 2016-07-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human chondroadherin: solving a difficult molecular-replacement problem using de novo models.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5LJV
 
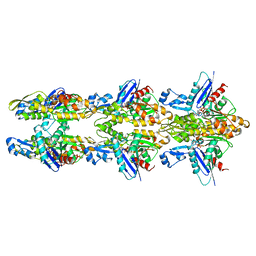 | | MamK double helical filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like ATPase, MAGNESIUM ION | | Authors: | Lowe, J. | | Deposit date: | 2016-07-21 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | X-ray and cryo-EM structures of monomeric and filamentous actin-like protein MamK reveal changes associated with polymerization.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1M8M
 
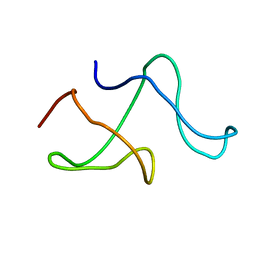 | | SOLID-STATE MAS NMR STRUCTURE OF THE A-SPECTRIN SH3 DOMAIN | | Descriptor: | SPECTRIN ALPHA CHAIN, BRAIN | | Authors: | Castellani, F, Van Rossum, B, Diehl, A, Schubert, M, Rehbein, K, Oschkinat, H. | | Deposit date: | 2002-07-25 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy
Nature, 420, 2002
|
|
6VH8
 
 | |
6FEB
 
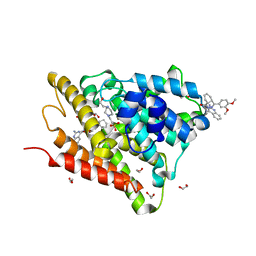 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-1086 | | Descriptor: | (4aS,8aR)-2-[1-(2-aminoquinazolin-4-yl)piperidin-4-yl]-4-(3,4-dimethoxyphenyl)-1,2,4a,5,8,8a-hexahydrophthalazin-1-one, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2017-12-31 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-1086
To be published
|
|
6MIE
 
 | | Solution NMR structure of the KCNQ1 voltage-sensing domain | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Taylor, K.C, Kuenze, G, Smith, J.A, Meiler, J, McFeeters, R.L, Sanders, C.R. | | Deposit date: | 2018-09-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and physiological function of the human KCNQ1 channel voltage sensor intermediate state.
Elife, 9, 2020
|
|
6FDI
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-226 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-[4-[(4~{a}~{S},8~{a}~{R})-4-(3,4-dimethoxyphenyl)-1-oxidanylidene-4~{a},5,8,8~{a}-tetrahydrophthalazin-2-yl]piperi din-1-yl]-2-oxidanylidene-ethyl]-4,4-dimethyl-piperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2017-12-24 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-226
To be published
|
|
6FW3
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-007 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{4-[(4aS,8aR)-4-(3,4-dimethoxyphenyl)-1-oxo-1,2,4a,5,8,8a-hexahydrophthalazin-2-yl]piperidin-1-yl}-2-oxoethyl)pyrrolidine-2,5-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-03-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-007
To be published
|
|
6FE7
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-356 | | Descriptor: | (4aS,8aR)-2-(1-{2-aminothieno[2,3-d]pyrimidin-4-yl}piperidin-4-yl)-4-(3,4- dimethoxyphenyl)-1,2,4a,5,8,8a-hexahydrophthalazin-1-one, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2017-12-29 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-356
To be published
|
|
6FET
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-1439 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{4-[(4aS,8aR)-4-[3,4-bis(difluoromethoxy)phenyl]-1-oxo-1,2,4a,5,8,8a-hexahydrophthalazin-2-yl]piperidin-1-yl}-2-oxoethyl)-4,4-dimethylpiperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-01-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-1439
To be published
|
|
6OGF
 
 | | 70S termination complex with RF2 bound to the UGA codon. Partially rotated ribosome with RF2 bound (Structure III). | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-02 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6FT0
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-425 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-{4-methoxy-3-[(2-methoxyphenyl)methoxy]phenyl}-2-(1-{thieno[3,2-d]pyrimidin-4-yl}piperidin-4-yl)-1,2,4a,5,8,8a-hexahydrophthalazin-1-one, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-02-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-425
To be published
|
|
