5UWX
 
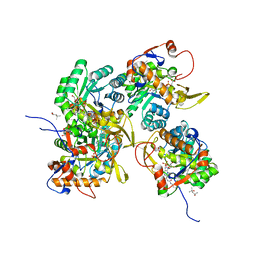 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P176 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-21 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P176
To Be Published
|
|
5UXE
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P178 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FORMIC ACID, INOSINIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P178
To Be Published
|
|
5ECR
 
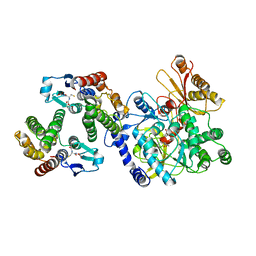 | | Crystal Structure of FIN219-FIP1 complex with JA, VAL and Mg | | Descriptor: | GLUTATHIONE, Glutathione S-transferase U20, Jasmonic acid-amido synthetase JAR1, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V5Y
 
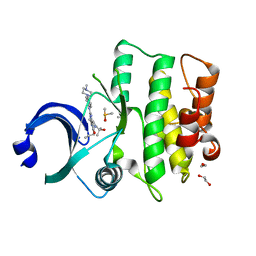 | | CRYSTAL STRUCTURE OF HUMAN WEE1 KINASE DOMAIN IN COMPLEX WITH MK1775 | | Descriptor: | 1,2-ETHANEDIOL, 1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | ZHU, J.-Y, SCHONBRUNN, E. | | Deposit date: | 2017-03-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
5DKJ
 
 | | Yeast 20S proteasome in complex with octreotide-PI | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Beck, P, Cui, H, Groll, M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeted Delivery of Proteasome Inhibitors to Somatostatin-Receptor-Expressing Cancer Cells by Octreotide Conjugation.
Chemmedchem, 10, 2015
|
|
5VD7
 
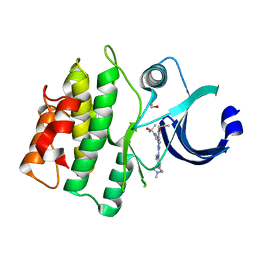 | | CRYSTAL STRUCTURE OF HUMAN WEE1 KINASE DOMAIN IN COMPLEX WITH RAC-IV-098, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 6-{[3-fluoro-4-(4-methylpiperazin-1-yl)phenyl]amino}-1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
5ECM
 
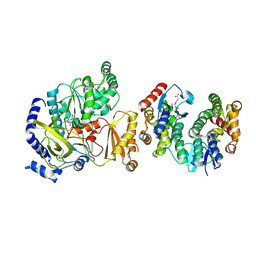 | | Crystal Structure of FIN219-FIP1 complex with JA and Leu | | Descriptor: | GLUTATHIONE, Glutathione S-transferase U20, Jasmonic acid-amido synthetase JAR1, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VD5
 
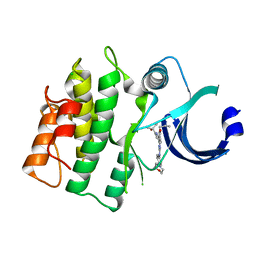 | |
5VDA
 
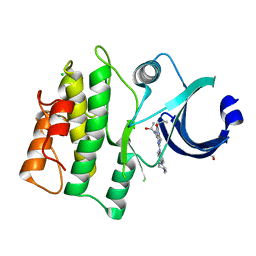 | | Crystal structure of human WEE1 kinase domain in complex with RAC-IV-101, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 1-{6-[(1S)-1-hydroxyethyl]pyridin-2-yl}-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
5ECO
 
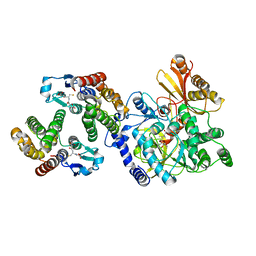 | | Crystal Structure of FIN219-FIP1 complex with JA, Leu and Mg | | Descriptor: | GLUTATHIONE, Glutathione S-transferase U20, Jasmonic acid-amido synthetase JAR1, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VD9
 
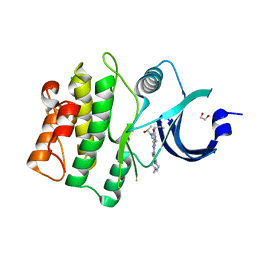 | | Crystal structure of human WEE1 kinase domain in complex with RAC-IV-097, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 1-{6-[(1R)-1-hydroxyethyl]pyridin-2-yl}-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
5ECH
 
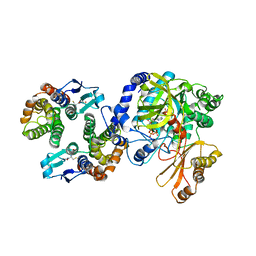 | | Crystal Structure of FIN219-FIP1 complex with JA and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE, Glutathione S-transferase U20, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W8U
 
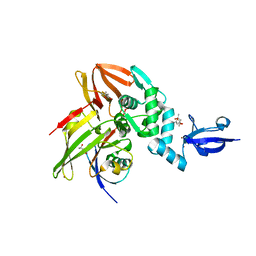 | | Crystal structure of MERS-CoV papain-like protease in complex with the C-terminal domain of human ISG15 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ORF1ab, Ubiquitin-like protein ISG15, ... | | Authors: | Daczkowski, C.M, Goodwin, O.Y, Dzimianski, J.D, Pegan, S.D. | | Deposit date: | 2017-06-22 | | Release date: | 2017-09-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structurally Guided Removal of DeISGylase Biochemical Activity from Papain-Like Protease Originating from Middle East Respiratory Syndrome Coronavirus.
J. Virol., 91, 2017
|
|
5WJ1
 
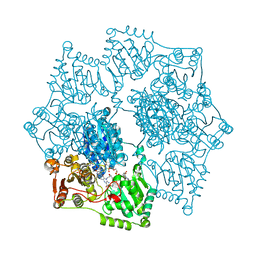 | | Crystal structure of Arabidopsis thaliana acetohydroxyacid synthase in complex with a triazolopyrimidine herbicide, penoxsulam | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 2-(2,2-difluoroethoxy)-N-(5,8-dimethoxy[1,2,4]triazolo[1,5-c]pyrimidin-2-yl)-6-(trifluoromethyl)benzenesulfonamide, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2017-07-21 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structural insights into the mechanism of inhibition of AHAS by herbicides.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5URS
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P178 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-12 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P178
To Be Published
|
|
5VD0
 
 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN IN COMPLEX WITH MK1775 | | Descriptor: | 1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, DI(HYDROXYETHYL)ETHER, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
5W7S
 
 | | Crystal structure of OxaC in complex with sinefungin and meleagrin | | Descriptor: | (3E,7aR,12aS)-6-hydroxy-3-[(1H-imidazol-4-yl)methylidene]-12-methoxy-7a-(2-methylbut-3-en-2-yl)-7a,12-dihydro-1H,5H-imidazo[1',2':1,2]pyrido[2,3-b]indole-2,5(3H)-dione, OxaC, SINEFUNGIN | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.948 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
5W8T
 
 | | Crystal structure of MERS-CoV papain-like protease in complex with the C-terminal domain of human ISG15 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ORF1ab, Ubiquitin-like protein ISG15, ... | | Authors: | Daczkowski, C.M, Goodwin, O.Y, Dzimianski, J.V, Farhat, J.J, Pegan, S.D. | | Deposit date: | 2017-06-22 | | Release date: | 2017-09-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Structurally Guided Removal of DeISGylase Biochemical Activity from Papain-Like Protease Originating from Middle East Respiratory Syndrome Coronavirus.
J. Virol., 91, 2017
|
|
5ECI
 
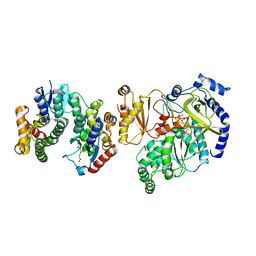 | | Crystal Structure of FIN219-FIP1 complex with JA, ATP and Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE, Glutathione S-transferase U20, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V84
 
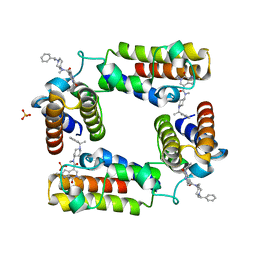 | | CECR2 in complex with Cpd6 (6-allyl-N,2-dimethyl-7-oxo-N-(1-(1-phenylethyl)piperidin-4-yl)-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-4-carboxamide) | | Descriptor: | Cat eye syndrome critical region protein 2, N,2-dimethyl-7-oxo-N-{1-[(1S)-1-phenylethyl]piperidin-4-yl}-6-(prop-2-en-1-yl)-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-4- carboxamide, SULFATE ION | | Authors: | Murray, J.M, Kiefer, J.R, Jayaran, H, Bellon, S, Boy, F. | | Deposit date: | 2017-03-21 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | GNE-886: A Potent and Selective Inhibitor of the Cat Eye Syndrome Chromosome Region Candidate 2 Bromodomain (CECR2).
ACS Med Chem Lett, 8, 2017
|
|
5WKC
 
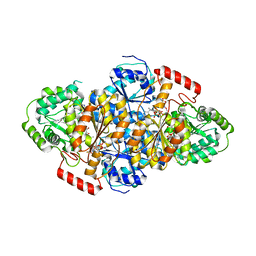 | | Saccharomyces cerevisiae acetohydroxyacid synthase in complex with the herbicide penoxsulam | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 2-(2,2-difluoroethoxy)-N-(5,8-dimethoxy[1,2,4]triazolo[1,5-c]pyrimidin-2-yl)-6-(trifluoromethyl)benzenesulfonamide, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, ... | | Authors: | Guddat, W.L, Lonhienne, G.T. | | Deposit date: | 2017-07-25 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structural insights into the mechanism of inhibition of AHAS by herbicides.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5UJV
 
 | | Crystal structure of FePYR1 in complex with abscisic acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, PYR1 | | Authors: | Ren, Z, Wang, Z, Zhou, X.E, Hong, Y, Cao, M, Chan, Z, Liu, X, Shi, H, Xu, H.E, Zhu, J.-K. | | Deposit date: | 2017-01-19 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure determination and activity manipulation of the turfgrass ABA receptor FePYR1.
Sci Rep, 7, 2017
|
|
6RLR
 
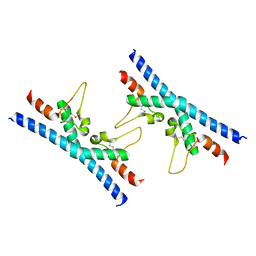 | | Crystal structure of CD9 large extracellular loop | | Descriptor: | CD9 antigen | | Authors: | Neviani, V, Kroon-Batenburg, L, Lutz, M, Pearce, N.M, Pos, W, Schotte, R, Spits, H, Gros, P. | | Deposit date: | 2019-05-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
5UQF
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with IMP and the inhibitor P225 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Campylobacter jejuni in the complex with IMP and the inhibitor P225
To Be Published
|
|
5URQ
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p176 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-{2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenyl}-alpha-D-ribofuranosylamine, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-12 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p176
To Be Published
|
|
