9II7
 
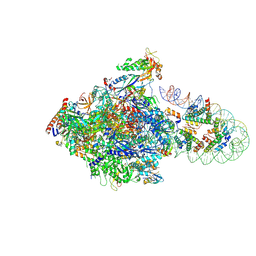 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome containing histone variant H2A.B | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Kujirai, T, Rina, H, Ehara, H, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2024-06-19 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of RNAPII transcription on the nucleosome containing histone variant H2A.B.
Embo J., 2025
|
|
4YM6
 
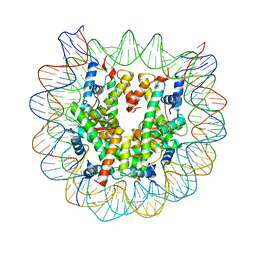 | | Crystal structure of the human nucleosome containing 6-4PP (outside) | | Descriptor: | 145-MER DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
9J0P
 
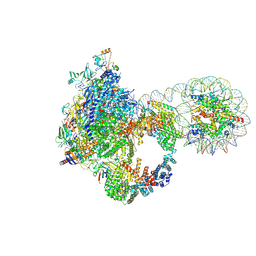 | | Arrested elongation complex of mammalian RNA polymerase II with nucleosome (AEC2-nuc) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Naganuma, M, Kujirai, T, Ehara, H, Uejima, T, Ito, T, Goto, M, Aoki, M, Henmi, M, Miyamoto-Kohno, S, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2024-08-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into promoter-proximal pausing of RNA polymerase II at +1 nucleosome.
Sci Adv, 11, 2025
|
|
9J0N
 
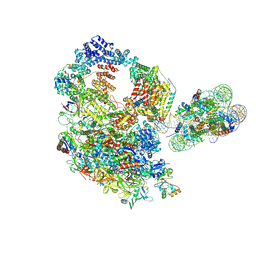 | | Paused elongation complex of mammalian RNA polymerase II with nucleosome (PEC2-nuc) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Naganuma, M, Kujirai, T, Ehara, H, Uejima, T, Ito, T, Goto, M, Aoki, M, Henmi, M, Miyamoto-Kohno, S, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2024-08-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into promoter-proximal pausing of RNA polymerase II at +1 nucleosome.
Sci Adv, 11, 2025
|
|
9J0O
 
 | | Arrested elongation complex of mammalian RNA polymerase II with nucleosome (AEC1-nuc) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Naganuma, M, Kujirai, T, Ehara, H, Uejima, T, Ito, T, Goto, M, Aoki, M, Henmi, M, Miyamoto-Kohno, S, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2024-08-02 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into promoter-proximal pausing of RNA polymerase II at +1 nucleosome.
Sci Adv, 11, 2025
|
|
4YM5
 
 | | Crystal structure of the human nucleosome containing 6-4PP (inside) | | Descriptor: | 144 mer-DNA, 144-mer DNA, Histone H2A type 1-B/E, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
7C0M
 
 | | Human cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Kujirai, T, Zierhut, C, Takizawa, Y, Kim, R, Negishi, L, Uruma, N, Hirai, S, Funabiki, H, Kurumizaka, H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for the inhibition of cGAS by nucleosomes.
Science, 370, 2020
|
|
9LJ2
 
 | | Structure of isw1-nucleosome double-bound complex in ADP-ADP+ state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2025-01-14 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
7COW
 
 | | 353 bp di-nucleosome harboring cohesive DNA termini with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (353-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-08-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
8VWT
 
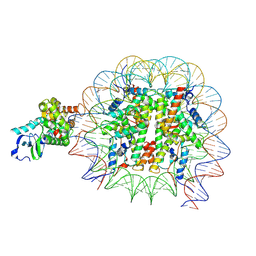 | | OGG1 bound to a nucleosome containing 8oxoG at SHL-6 (composite map) | | Descriptor: | 601 I strand (non-damaged strand), 601 J strand (damaged strand), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
8VWU
 
 | | Nucleosome containing 8oxoG at SHL4 | | Descriptor: | 601 I strand (damaged strand), 601 J strand (non-damaged strand), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
8VWV
 
 | | OGG1 bound to a nucleosome containing 8oxoG at SHL4 (composite map) | | Descriptor: | 601 I strand (damaged strand), 601 J strand (non-damaged), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
8VWS
 
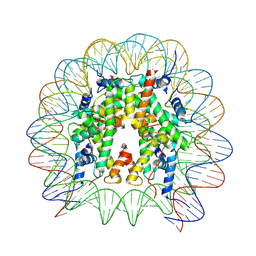 | | Nucleosome containing 8oxoG at SHL-6 | | Descriptor: | 601 I strand (non-damaged strand), 601 J strand (damaged strand), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
8XJV
 
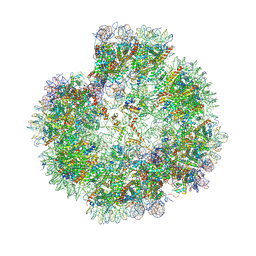 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
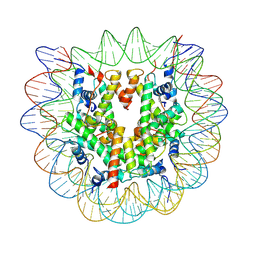
7U51
 
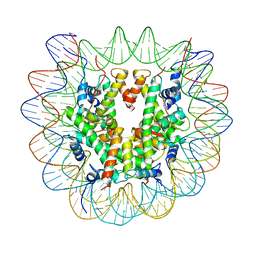 | |
7U52
 
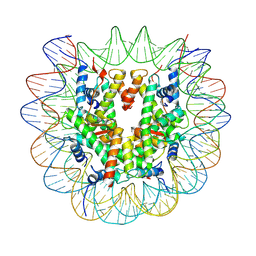 | |
7U50
 
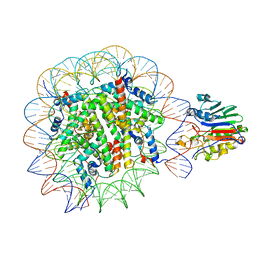 | |
7U53
 
 | |
7YQK
 
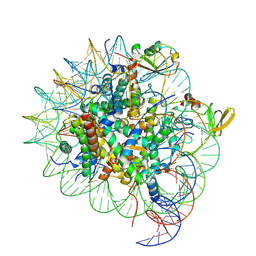 | | cryo-EM structure of gammaH2AXK15ub-H4K20me2 nucleosome bound to 53BP1 | | Descriptor: | DNA (145-MER), Histone H2AX, Histone H2B, ... | | Authors: | Ai, H.S, GuoChao, C, Qingyue, G, Ze-Bin, T, Zhiheng, D, Xin, L, Fan, Y, Ziyu, X, Jia-Bin, L, Changlin, T, Liu, L. | | Deposit date: | 2022-08-07 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Chemical Synthesis of Post-Translationally Modified H2AX Reveals Redundancy in Interplay between Histone Phosphorylation, Ubiquitination, and Methylation on the Binding of 53BP1 with Nucleosomes.
J.Am.Chem.Soc., 144, 2022
|
|
8ZVY
 
 | | Alpha-Synuclein with H2a-H2b dimer complex structure. | | Descriptor: | CHLORIDE ION, Histone H2B 1.1,Histone H2A type 1, Isoform 1 of Alpha-synuclein | | Authors: | Padavattan, S, Jos, S, Kambaru, A. | | Deposit date: | 2024-06-12 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and functional insights into the nuclear role of Parkinson's disease-associated alpha-synuclein as a histone chaperone.
Commun Biol, 8, 2025
|
|
7LBP
 
 | |
6WAV
 
 | | Crystal structure of PHF1 in complex with H3K36me3 substitution | | Descriptor: | Histone H3.1, PHD finger protein 1, SULFATE ION, ... | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
6V41
 
 | | crystal structure of CDY1 chromodomain bound to H3K9me3 | | Descriptor: | Histone H3.1 Peptide, Testis-specific chromodomain protein Y 1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-27 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
9DWK
 
 | |
9DWF
 
 | |
