8CVJ
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-NH-tRNAphe, peptidyl P-site fMSEAC-NH-tRNAmet, and deacylated E-site tRNAphe at 2.40A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | Deposit date: | 2022-05-18 | | Release date: | 2022-10-19 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the ribosome function from the structures of non-arrested ribosome-nascent chain complexes.
Nat.Chem., 15, 2023
|
|
8CVK
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-NH-tRNAphe, peptidyl P-site fMRC-NH-tRNAmet, and deacylated E-site tRNAphe at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | Deposit date: | 2022-05-18 | | Release date: | 2022-10-19 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the ribosome function from the structures of non-arrested ribosome-nascent chain complexes.
Nat.Chem., 15, 2023
|
|
8CVL
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-NH-tRNAphe, peptidyl P-site fMTHSMRC-NH-tRNAmet, and deacylated E-site tRNAphe at 2.30A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | Deposit date: | 2022-05-18 | | Release date: | 2022-10-19 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into the ribosome function from the structures of non-arrested ribosome-nascent chain complexes.
Nat.Chem., 15, 2023
|
|
6ZQM
 
 | | bovine ATP synthase monomer state 2 (combined) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-10 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZJ3
 
 | | Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis | | Descriptor: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Matzov, D, Halfon, H, Zimmerman, E, Rozenberg, H, Bashan, A, Gray, M.W, Yonath, A.E, Shalev-Benami, M. | | Deposit date: | 2020-06-27 | | Release date: | 2020-10-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis.
Nucleic Acids Res., 48, 2020
|
|
8B7A
 
 | | Tubulin - maytansinoid - 4 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Boiarska, Z, Perez-Pena, H, Abel, A.-C, Marzullo, P, Alvarez-Bernad, B, Bonato, F, Santini, B, Horvath, D, Lucena-Agell, D, Vasile, F, Sironi, M, Diaz, F.J, Steinmetz, M.O, Prota, A.E, Piaraccini, S, Passarella, D. | | Deposit date: | 2022-09-29 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Functionalization: Towards Useful Probes for Studying Microtubule Dynamics.
Chemistry, 29, 2023
|
|
8B7C
 
 | | Tubulin-maytansinoid-12 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Boiarska, Z, Perez-Pena, H, Abel, A.-C, Marzullo, P, Alvarez-Bernad, B, Bonato, F, Santini, B, Horvath, D, Lucena-Agell, D, Vasile, F, Sironi, M, Diaz, J.F, Steinmetz, M.O, Prota, A.E, Pieraccini, S, Passarella, D. | | Deposit date: | 2022-09-29 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Maytansinol Functionalization: Towards Useful Probes for Studying Microtubule Dynamics.
Chemistry, 29, 2023
|
|
6ZNA
 
 | | Porcine ATP synthase Fo domain | | Descriptor: | ATP synthase F(0) complex subunit C1, mitochondrial, ATP synthase g subunit, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-06 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8TJH
 
 | |
7AIW
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with (E)-10-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)-6-decenamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
6FKJ
 
 | | Tubulin-TUB075 complex | | Descriptor: | (5~{S})-2-[(~{E})-~{N}-(2-ethoxyphenyl)-~{C}-methyl-carbonimidoyl]-3-oxidanyl-5-phenyl-cyclohex-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Steinmetz, M.O, Priego, E.-M. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | High-affinity ligands of the colchicine domain in tubulin based on a structure-guided design.
Sci Rep, 8, 2018
|
|
6QE0
 
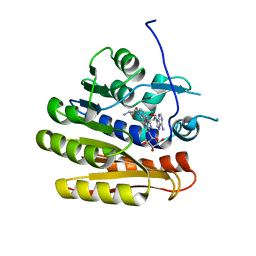 | | Structure of E.coli RlmJ in complex with a bisubstrate analogue (BA2) | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[2-[[9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-6-yl]amino]ethyl]amino]-2-azanyl-butanoic acid, Ribosomal RNA large subunit methyltransferase J | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-03 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.394 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
6GZ5
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-3 (TI-POST-3) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
3FFZ
 
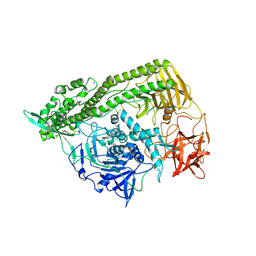 | | Domain organization in Clostridium butulinum neurotoxin type E is unique: Its implication in faster translocation | | Descriptor: | ACETATE ION, Botulinum neurotoxin type E, SODIUM ION, ... | | Authors: | Kumaran, D, Eswaramoorthy, S, Swaminathan, S. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Domain organization in Clostridium botulinum neurotoxin type E is unique: its implication in faster translocation.
J.Mol.Biol., 386, 2009
|
|
6NU9
 
 | |
8F3T
 
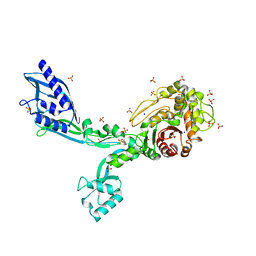 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SODIUM ION, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3H
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S466 insertion variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3L
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3S
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3O
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) R464A variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Peti, W, Page, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3N
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant with S466 insertion penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3U
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3F
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Hunashal, Y, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3R
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3P
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) R464A variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
