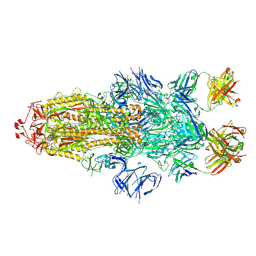
7MW2
 
 | |
3L5S
 
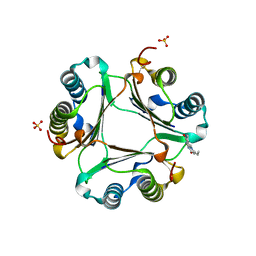 | |
6QYN
 
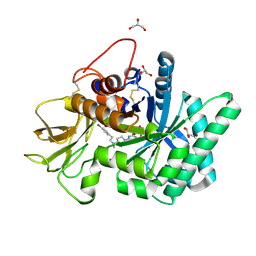
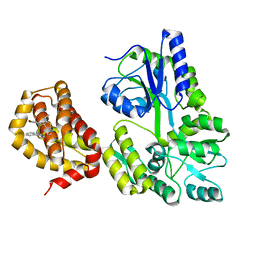 | | Structure of MBP-Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
5NR8
 
 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7a | | Descriptor: | 1-(3-azanyl-1~{H}-1,2,4-triazol-5-yl)-~{N}-[2-(4-bromophenyl)ethyl]-~{N}-methyl-piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | Authors: | Podjarny, A, Fadel, F, Golebiowski, A. | | Deposit date: | 2017-04-22 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
3LGC
 
 | | Crystal Structure of Glutaredoxin 1 from Francisella tularensis | | Descriptor: | GLYCEROL, Glutaredoxin 1, SULFATE ION | | Authors: | Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal Structure of Glutaredoxin 1 from Francisella tularensis
To be Published
|
|
7S05
 
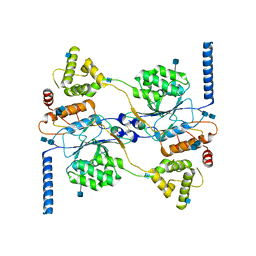 | | Cryo-EM structure of human GlcNAc-1-phosphotransferase A2B2 subcomplex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-08-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human GlcNAc-1-phosphotransferase alpha beta subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S06
 
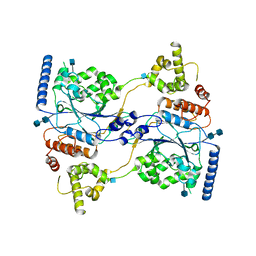 | | Cryo-EM structure of human GlcNAc-1-phosphotransferase A2B2 subcomplex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosamine-1-phosphotransferase subunits alpha/beta | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-08-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human GlcNAc-1-phosphotransferase alpha beta subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7U4O
 
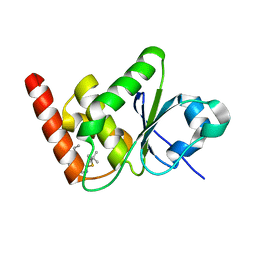 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-propyl-5,6-dihydrothieno[3,4-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 3,3-dimethyl-1-{[(9aM)-9-propyl-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, Dual specificity protein phosphatase 10 | | Authors: | Gannam, Z.T.K, Jamali, H, Lolis, E, Anderson, K.S, Ellman, J.A, Bennett, A.M. | | Deposit date: | 2022-02-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Defining the structure-activity relationship for a novel class of allosteric MKP5 inhibitors.
Eur.J.Med.Chem., 243, 2022
|
|
7T7N
 
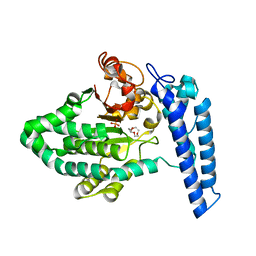 | | Structure of SPCC1393.13 protein from fission yeast | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Damage-control phosphatase SPCC1393.13, PHOSPHATE ION | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
7TE3
 
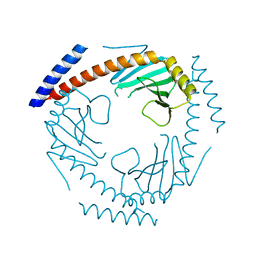 | | Crystal Structure of a Double Loop Deletion Mutant in gC1qR/C1qBP/HABP-1 | | Descriptor: | Complement component 1 Q subcomponent-binding protein, mitochondrial | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2022-01-04 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | gC1qR/C1qBP/HABP-1: Structural Analysis of the Trimeric Core Region, Interactions With a Novel Panel of Monoclonal Antibodies, and Their Influence on Binding to FXII.
Front Immunol, 13, 2022
|
|
6U4Z
 
 | |
5G1O
 
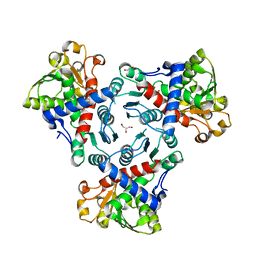 | | Aspartate transcarbamoylase domain of human CAD in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAD protein, GLYCEROL | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
7MU6
 
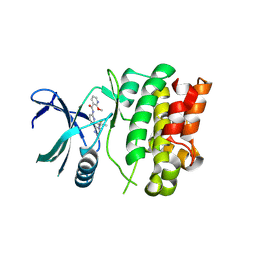 | | Ask1 bound to compound 28 | | Descriptor: | 2-methoxy-N-(6-{4-[(2S)-1,1,1-trifluoropropan-2-yl]-4H-1,2,4-triazol-3-yl}pyridin-2-yl)pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Chodaparambil, J.V, Marcotte, D.J. | | Deposit date: | 2021-05-14 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | Discovery of Potent, Selective, and Brain-Penetrant Apoptosis Signal-Regulating Kinase 1 (ASK1) Inhibitors that Modulate Brain Inflammation In Vivo.
J.Med.Chem., 64, 2021
|
|
7R8U
 
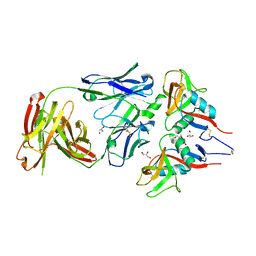 | |
6T6I
 
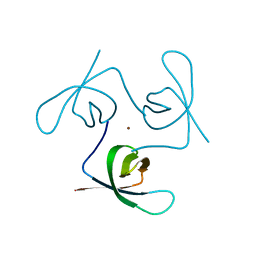 | |
8P2M
 
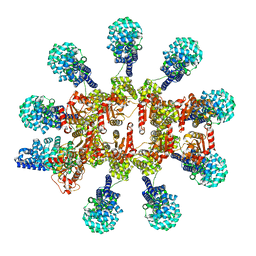 | | C. elegans TIR-1 protein. | | Descriptor: | NAD(+) hydrolase tir-1 | | Authors: | Isupov, M.N, Opatowsky, Y. | | Deposit date: | 2023-05-16 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure-function analysis of ceTIR-1/hSARM1 explains the lack of Wallerian axonal degeneration in C. elegans.
Cell Rep, 42, 2023
|
|
7RE5
 
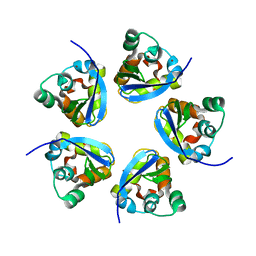 | |
8IAV
 
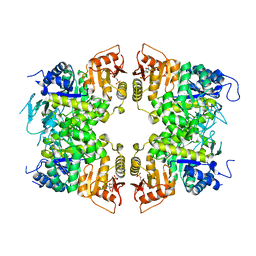 | |
8IAU
 
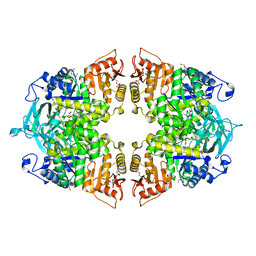 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with oxalate and fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of Streptococcus pneumoniae pyruvate kinase involved in fosfomycin resistance.
J.Biol.Chem., 299, 2023
|
|
6T6J
 
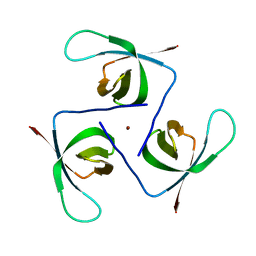 | |
7RRE
 
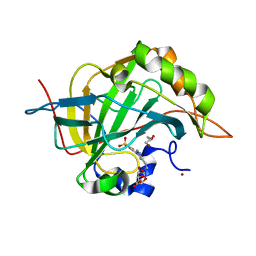 | | Carbonic Anhydrase II in complex with Beta-Galactose-2C | | Descriptor: | 2-[1-(1,1,3-trioxo-2,3-dihydro-1H-1lambda~6~,2-benzothiazol-6-yl)-1H-1,2,3-triazol-4-yl]ethyl beta-L-gulopyranoside, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | McKenna, R, Combs, J.E. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Inhibition of Carbonic Anhydrase IX and Monocarboxylate Transporters 1 and 4 in breast cancer via novel inhibition with Beta-Galactose 2C
To Be Published
|
|
7XIM
 
 | |
7BNT
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with a predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Descriptor: | 1,2-ETHANEDIOL, AVR-Pik protein, Predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Authors: | Bialas, A, Langner, T, Harant, A, Contreras, M.P, Stevenson, C.E.M, Lawson, D.M, Sklenar, J, Kellner, R, Moscou, M.J, Terauchi, R, Banfield, M.J, Kamoun, S. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Two NLR immune receptors acquired high-affinity binding to a fungal effector through convergent evolution of their integrated domain.
Elife, 10, 2021
|
|
8P1B
 
 | | Lysozyme structure solved from serial crystallography data collected at 2 kHz with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
7ZQ1
 
 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2205 | | Descriptor: | 1-[(8R,15S,18S)-15-(4-azanylbutyl)-18-(1H-indol-3-ylmethyl)-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-04-29 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Synthesis and structural characterization of new macrocyclic inhibitors of the Zika virus NS2B-NS3 protease.
Arch Pharm, 2024
|
|
