6KYA
 
 | | Crystal structure of human TLR8 in complex TH1027 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-methylsulfanyl-4-(3,4,5-trimethylphenyl)-1,2,4-triazol-3-yl]propan-1-ol, ... | | Authors: | Tanji, H, Sakaniwa, K, Ohto, U, Shimizu, T. | | Deposit date: | 2019-09-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Rationally Designed Small-Molecule Inhibitors Targeting an Unconventional Pocket on the TLR8 Protein-Protein Interface.
J.Med.Chem., 63, 2020
|
|
2M0N
 
 | |
3CXO
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and 3-deoxy-L-rhamnonate | | Descriptor: | (2R,4S)-2,4,7-trihydroxyheptanoic acid, 3,6-dideoxy-L-arabino-hexonic acid, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Hubbard, B.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-04-24 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase.
Biochemistry, 47, 2008
|
|
3DLP
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, Mutant D402P, bound to 4CB | | Descriptor: | 4-CHLORO-BENZOIC ACID, 4-Chlorobenzoate CoA Ligase/Synthetase | | Authors: | Wu, R, Cao, J, Reger, A.S, Lu, X, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2008-06-28 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The mechanism of domain alternation in the acyl-adenylate forming ligase superfamily member 4-chlorobenzoate: coenzyme A ligase
Biochemistry, 48, 2009
|
|
2W8S
 
 | | CRYSTAL STRUCTURE OF A catalytically promiscuous PHOSPHONATE MONOESTER HYDROLASE FROM Burkholderia caryophylli | | Descriptor: | FE (III) ION, GLYCEROL, PHOSPHONATE MONOESTER HYDROLASE, ... | | Authors: | Jonas, S, van Loo, B, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2009-01-19 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Efficient, Multiply Promiscuous Hydrolase in the Alkaline Phosphatase Superfamily.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7TA2
 
 | | Crystal structure of the human sperm-expressed surface protein SPACA6 | | Descriptor: | BROMIDE ION, Sperm acrosome membrane-associated protein 6 | | Authors: | Vance, T.D.R, Yip, P, Jimenez, E, Uson, I, Lee, J.E. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SPACA6 ectodomain structure reveals a conserved superfamily of gamete fusion-associated proteins.
Commun Biol, 5, 2022
|
|
6S3F
 
 | | Moringa seed protein Mo-CBP3-4 | | Descriptor: | 2S albumin, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Moulin, M, Mossou, E, Mitchell, E.P, Haertlein, M, Forsyth, V.T, Rennie, A.R. | | Deposit date: | 2019-06-25 | | Release date: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Towards a molecular understanding of the water purification properties of Moringa seed proteins.
J Colloid Interface Sci, 554, 2019
|
|
8IK0
 
 | |
2VNR
 
 | | Family 51 carbohydrate binding module from a family 98 glycoside hydrolase produced by Clostridium perfringens. | | Descriptor: | CALCIUM ION, CPE0329 | | Authors: | Gregg, K.J, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-02-06 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
4LRU
 
 | | Crystal structure of glyoxalase III (Orf 19.251) from Candida albicans | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glyoxalase III (glutathione-independent) | | Authors: | Hasim, S, Hussin, N.A, Nickerson, K.W, Wilson, M.A. | | Deposit date: | 2013-07-20 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Glutathione-independent Glyoxalase of the DJ-1 Superfamily Plays an Important Role in Managing Metabolically Generated Methylglyoxal in Candida albicans.
J.Biol.Chem., 289, 2014
|
|
3BOX
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg | | Descriptor: | L-rhamnonate dehydratase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase.
Biochemistry, 47, 2008
|
|
6FXF
 
 | |
4UPI
 
 | | Dimeric sulfatase SpAS1 from Silicibacter pomeroyi | | Descriptor: | SULFATASE FAMILY PROTEIN, ZINC ION | | Authors: | Jonas, S, van Loo, B, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Balancing Specificity and Promiscuity in Enzyme Evolution: Multidimensional Activity Transitions in the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 141, 2019
|
|
6G8O
 
 | |
2I5Q
 
 | | Crystal structure of Apo L-rhamnonate dehydratase from Escherichia Coli | | Descriptor: | L-rhamnonate dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase.
Biochemistry, 47, 2008
|
|
2VNO
 
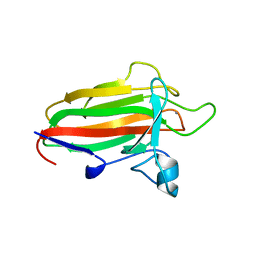 | | Family 51 carbohydrate binding module from a family 98 glycoside hydrolase produced by Clostridium perfringens in complex with blood group B-trisaccharide ligand. | | Descriptor: | CALCIUM ION, CPE0329, alpha-L-fucopyranose-(1-2)-[beta-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Gregg, K.J, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-02-05 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
7NPG
 
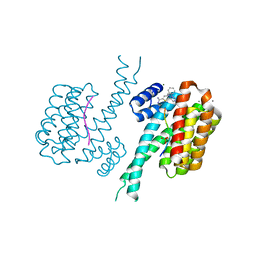 | |
3ES8
 
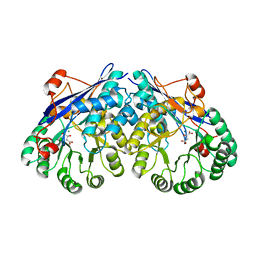 | | Crystal structure of divergent enolase from Oceanobacillus Iheyensis complexed with Mg and L-malate. | | Descriptor: | (2S)-2-hydroxybutanedioic acid, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-04 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
7OB3
 
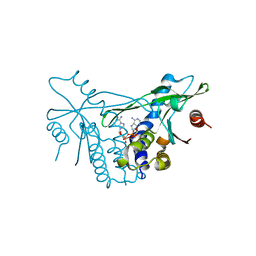 | | hSTING in complex with 3',3'-c-di-araAMP | | Descriptor: | 3',3'-c-di-araAMP, Stimulator of interferon genes protein | | Authors: | Smola, M, Boura, E. | | Deposit date: | 2021-04-20 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Synthesis of 3'-5', 3'-5' Cyclic Dinucleotides, Their Binding Properties to the Stimulator of Interferon Genes Adaptor Protein, and Structure/Activity Correlations
Biochemistry, 2021
|
|
3ES7
 
 | | Crystal structure of divergent enolase from Oceanobacillus Iheyensis complexed with Mg and L-malate. | | Descriptor: | (2S)-2-hydroxybutanedioic acid, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-04 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
6O47
 
 | | human cGAS core domain (K427E/K428E) bound with RU-521 | | Descriptor: | (3~{S})-3-[1-[4,5-bis(chloranyl)-1~{H}-benzimidazol-2-yl]-3-methyl-5-oxidanyl-pyrazol-4-yl]-3~{H}-2-benzofuran-1-one, 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, CITRIC ACID, ... | | Authors: | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1WR8
 
 | |
6EXU
 
 | |
6EXT
 
 | |
4Q2C
 
 | | Crystal structure of CRISPR-associated protein | | Descriptor: | CRISPR-associated helicase Cas3, NICKEL (II) ION | | Authors: | Gong, B, Shin, M, Sun, J, van der Oost, J, Kim, J.-S. | | Deposit date: | 2014-04-07 | | Release date: | 2014-11-19 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
