4G21
 
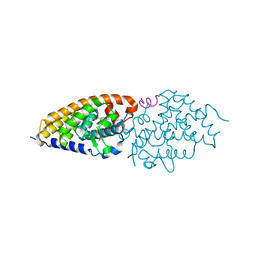 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-ethyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4TTM
 
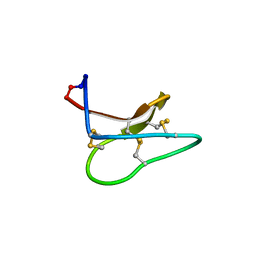 | | Racemic structure of kalata B1 (kB1) | | Descriptor: | D-kalata B1, Kalata-B1 | | Authors: | Wang, C.K, King, G.J, Craik, D.J. | | Deposit date: | 2014-06-22 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9001 Å) | | Cite: | Racemic and Quasi-Racemic X-ray Structures of Cyclic Disulfide-Rich Peptide Drug Scaffolds.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4AG9
 
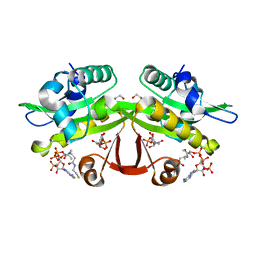 | | C. elegans glucosamine-6-phosphate N-acetyltransferase (GNA1): ternary complex with coenzyme A and GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, COENZYME A, ... | | Authors: | Dorfmueller, H.C, Fang, W, Rao, F.V, Blair, D.E, Attrill, H, Shepherd, S.M, van Aalten, D.M.F. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Biochemical Characterization of a Trapped Coenzyme a Adduct of Caenorhabditis Elegans Glucosamine-6-Phosphate N-Acetyltransferase 1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1NZC
 
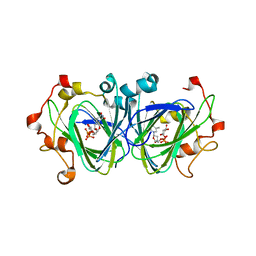 | | The high resolution structures of RmlC from Streptococcus suis in complex with dTDP-D-xylose | | Descriptor: | NICKEL (II) ION, THYMIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE, dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | Authors: | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | Deposit date: | 2003-02-17 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
2BES
 
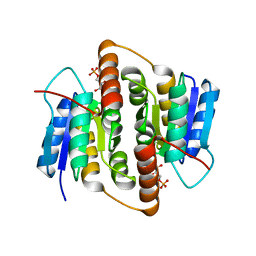 | | Structure of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, in complex with 4-phospho-D-erythronohydroxamic acid. | | Descriptor: | 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, CARBOHYDRATE-PHOSPHATE ISOMERASE | | Authors: | Roos, A.K, Ericsson, D.J, Mowbray, S.L. | | Deposit date: | 2004-11-30 | | Release date: | 2004-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive Inhibitors of Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase B Reveal New Information About the Reaction Mechanism.
J.Biol.Chem., 280, 2005
|
|
4PF8
 
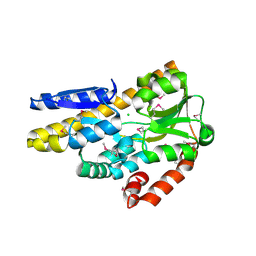 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFITOBACTER sp. NAS-14.1 (TARGET EFI-510299) WITH BOUND BETA-D-GALACTURONATE | | Descriptor: | CHLORIDE ION, TRAP-T family transporter, DctP (Periplasmic binding) subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
2Q2R
 
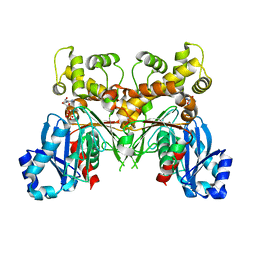 | | Trypanosoma cruzi glucokinase in complex with beta-D-glucose and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glucokinase 1, putative, ... | | Authors: | Cordeiro, A.T, Caceres, A.J, Vertommen, D, Concepcion, J.L, Michels, P.A, Versees, W. | | Deposit date: | 2007-05-29 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Trypanosoma cruzi Glucokinase Reveals Features Determining Oligomerization and Anomer Specificity of Hexose-phosphorylating Enzymes.
J.Mol.Biol., 372, 2007
|
|
4OIA
 
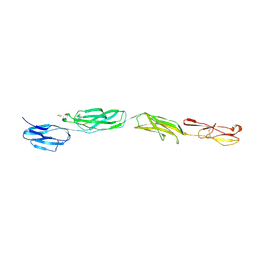 | | Crystal Structure of ICAM-5 D1-D4 ectodomain fragment, Space Group P4322 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Recacha, R, Jimenez, D, Tian, L, Barredo, R, Ghamberg, C, Casasnovas, J.M. | | Deposit date: | 2014-01-19 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structures of an ICAM-5 ectodomain fragment show electrostatic-based homophilic adhesions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4KCU
 
 | | Pyruvate kinase (PYK) from Trypanosoma brucei soaked with D-Malate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, D-MALATE, MAGNESIUM ION, ... | | Authors: | Zhong, W, Morgan, H.P, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2013-04-24 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pyruvate kinases have an intrinsic and conserved decarboxylase activity.
Biochem.J., 458, 2014
|
|
2BET
 
 | | Structure of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, in complex with 4-phospho-D-erythronate. | | Descriptor: | 4-PHOSPHO-D-ERYTHRONATE, CARBOHYDRATE-PHOSPHATE ISOMERASE | | Authors: | Roos, A.K, Ericsson, D.J, Mowbray, S.L. | | Deposit date: | 2004-11-30 | | Release date: | 2004-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Competitive Inhibitors of Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase B Reveal New Information About the Reaction Mechanism
J.Biol.Chem., 280, 2005
|
|
4G2U
 
 | | Crystal Structure Analysis of Ostertagia ostertagi ASP-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ancylostoma-secreted protein-like protein, SULFATE ION | | Authors: | Weeks, S.D, Borloo, J, Geldhof, P, Vercruysse, J, Strelkov, S.V. | | Deposit date: | 2012-07-13 | | Release date: | 2013-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Ostertagia ostertagi ASP-1: insights into disulfide-mediated cyclization and dimerization
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1Q0Q
 
 | | Crystal structure of DXR in complex with the substrate 1-deoxy-D-xylulose-5-phosphate | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | Deposit date: | 2003-07-17 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
7EC3
 
 | | Crystal structure of SdgB (complexed with UDP, GlcNAc, and Glycosylated peptide) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-35)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-65)]5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyl transferase, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3C2U
 
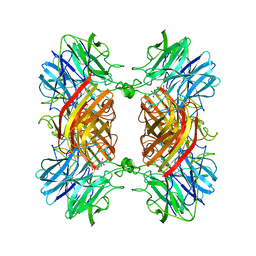 | | Structure of the two subsite D-xylosidase from Selenomonas ruminantium in complex with 1,3-bis[tris(hydroxymethyl)methylamino]propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Xylosidase/arabinosidase | | Authors: | Brunzelle, J.S, Jordan, D.B, McCaslin, D.R, Olczak, A, Wawrzak, A. | | Deposit date: | 2008-01-25 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the two-subsite beta-d-xylosidase from Selenomonas ruminantium in complex with 1,3-bis[tris(hydroxymethyl)methylamino]propane.
Arch.Biochem.Biophys., 474, 2008
|
|
4GNV
 
 | |
1EEH
 
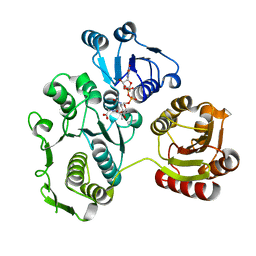 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J.A, Fanchon, E, Martin, L, Chantalat, L, Auger, G, Blanot, D, van Heijenoort, J, Dideberg, O. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | "Open" structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase.
J.Mol.Biol., 301, 2000
|
|
1OJ8
 
 | | Novel and retro Binding Modes in Cytotoxic Ribonucleases from Rana catesbeiana of Two Crystal Structures Complexed with d(ApCpGpA) and (2',5'CpG) | | Descriptor: | 5'-D(*AP*CP*GP*AP)-3', RC-RNASE6 RIBONUCLEASE, SULFATE ION | | Authors: | Tsai, C.-J, Liu, J.-H, Liao, Y.-D, Chen, L.-Y, Cheng, P.-T, Sun, Y.-J. | | Deposit date: | 2003-07-07 | | Release date: | 2004-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Retro and Novel Binding Modes in Cytotoxic Ribonucleases from Rana Catesbeiana of Two Crystal Structures Complexed with (2',5'Cpg) and D(Apcpgpa)
To be Published
|
|
1DA9
 
 | | ANTHRACYCLINE-DNA INTERACTIONS AT UNFAVOURABLE BASE BASE-PAIR TRIPLET-BINDING SITES: STRUCTURES OF D(CGGCCG)/DAUNOMYCIN AND D(TGGCCA)/ADRIAMYCIN COMPL | | Descriptor: | DNA (5'-D(*TP*GP*GP*CP*CP*A)-3'), DOXORUBICIN | | Authors: | Leonard, G.A, Hambley, T.W, McAuley-Hecht, K, Brown, T, Hunter, W.N. | | Deposit date: | 1993-01-21 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anthracycline-DNA interactions at unfavourable base-pair triplet-binding sites: structures of d(CGGCCG)/daunomycin and d(TGGCCA)/adriamycin complexes.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1P4J
 
 | | Crystal structure of glycogen phosphorylase b in complex with C-(1-hydroxy-beta-D-glucopyranosyl)formamide | | Descriptor: | (2R,3R,4S,5S,6R)-2,3,4,5-tetrahydroxy-6-(hydroxymethyl)oxane-2-carboxamide, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Oikonomakos, N.G, Zographos, S.E, Kosmopoulou, M.N, Bischler, N, Leonidas, D.D, Nagy, V, Somsak, L, Gergely, P, Praly, J.-P. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crsytallographic Studies on alpha- and beta-D-glucopyranosyl Formamide Analogues, Inhibitors of Glycogen Phosphorylase
Biocatal.Biotransfor., 21, 2003
|
|
4D9Q
 
 | |
4D9R
 
 | |
2NPW
 
 | | Solution Structures of a DNA Dodecamer Duplex with a Cisplatin 1,2-d(GG) Intrastrand Cross-Link | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', Cisplatin | | Authors: | Wu, Y, Bhattacharyya, D, Chaney, S, Campbell, S. | | Deposit date: | 2006-10-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a DNA Dodecamer Duplex with and without a Cisplatin 1,2-d(GG) Intrastrand Cross-Link: Comparison with the Same DNA Duplex Containing an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link
Biochemistry, 46, 2007
|
|
2VAR
 
 | | Crystal structure of Sulfolobus solfataricus 2-keto-3-deoxygluconate kinase complexed with 2-keto-3-deoxygluconate | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, 3-deoxy-alpha-D-erythro-hex-2-ulofuranosonic acid, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Potter, J.A, Theodossis, A, Kerou, M, Lamble, H.J, Bull, S.D, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2007-09-04 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Sulfolobus Solfataricus 2-Keto-3-Deoxygluconate Kinase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2NQ0
 
 | | Solution Structures of a DNA Dodecamer Duplex with a Cisplatin 1,2-d(GG) Intrastrand Cross-Link | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', Cisplatin | | Authors: | Wu, Y, Bhattacharyya, D, Chaney, S, Campbell, S. | | Deposit date: | 2006-10-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a DNA Dodecamer Duplex with and without a Cisplatin 1,2-d(GG) Intrastrand Cross-Link: Comparison with the Same DNA Duplex Containing an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link
Biochemistry, 46, 2007
|
|
1LMO
 
 | |
