5IH5
 
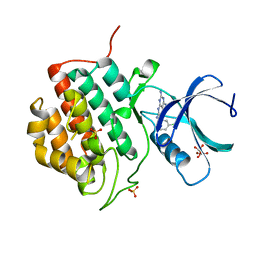 | | Human Casein Kinase 1 isoform delta (kinase domain) in complex with Epiblastin A | | Descriptor: | 6-(3-chlorophenyl)pteridine-2,4,7-triamine, Casein kinase I isoform delta, S,R MESO-TARTARIC ACID, ... | | Authors: | Ursu, A, Illich, D.J, Takemoto, Y, Porfetye, A.T, Zhang, M, Brockmeyer, A, Janning, P, Watanabe, N, Osada, H, Vetter, I.R, Ziegler, S, Schoeler, H.R, Waldmann, H. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epiblastin A Induces Reprogramming of Epiblast Stem Cells Into Embryonic Stem Cells by Inhibition of Casein Kinase 1.
Cell Chem Biol, 23, 2016
|
|
5IH6
 
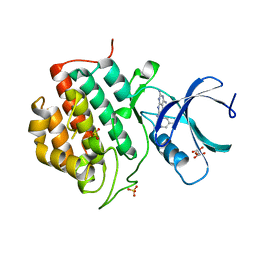 | | Human Casein Kinase 1 isoform delta (kinase domain) in complex with Epiblastin A derivative | | Descriptor: | 6-(3-bromophenyl)pteridine-2,4,7-triamine, Casein kinase I isoform delta, S,R MESO-TARTARIC ACID, ... | | Authors: | Ursu, A, Illich, D.J, Takemoto, Y, Porfetye, A.T, Zhang, M, Brockmeyer, A, Janning, P, Watanabe, N, Osada, H, Vetter, I.R, Ziegler, S, Schoeler, H.R, Waldmann, H. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Epiblastin A Induces Reprogramming of Epiblast Stem Cells Into Embryonic Stem Cells by Inhibition of Casein Kinase 1.
Cell Chem Biol, 23, 2016
|
|
5IH4
 
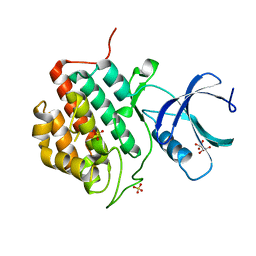 | | Human Casein Kinase 1 isoform delta apo (kinase domain) | | Descriptor: | Casein kinase I isoform delta, S,R MESO-TARTARIC ACID, SULFATE ION, ... | | Authors: | Ursu, A, Illich, D.J, Takemoto, Y, Porfetye, A.T, Zhang, M, Brockmeyer, A, Janning, P, Watanabe, N, Osada, H, Vetter, I.R, Ziegler, S, Schoeler, H.R, Waldmann, H. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Epiblastin A Induces Reprogramming of Epiblast Stem Cells Into Embryonic Stem Cells by Inhibition of Casein Kinase 1.
Cell Chem Biol, 23, 2016
|
|
5JG4
 
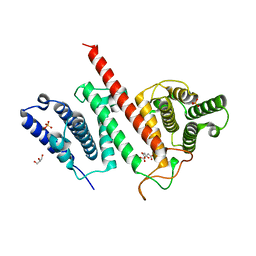 | | Structure of the effector protein LpiR1 (Lpg0634) from Legionella pneumophila | | Descriptor: | CITRATE ANION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Beyrakhova, K, van Straaten, K, Cygler, M. | | Deposit date: | 2016-04-19 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Investigations of the Effector Protein LpiR1 from Legionella pneumophila.
J.Biol.Chem., 291, 2016
|
|
4PQH
 
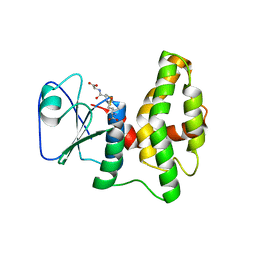 | | Crystal structure of glutathione transferase lambda1 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, SODIUM ION, glutathione transferase lambda1 | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
5XV3
 
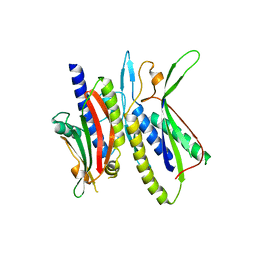 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13, DI(HYDROXYETHYL)ETHER | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5XV6
 
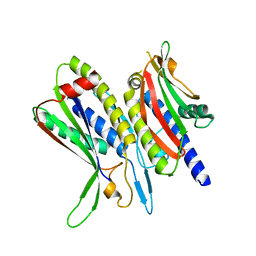 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
2UYF
 
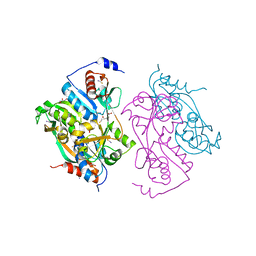 | | Single mutant F111L DntR from Burkholderia sp. strain DNT in complex with thiocyanate | | Descriptor: | ACETATE ION, GLYCEROL, REGULATORY PROTEIN, ... | | Authors: | Lonneborg, R, Smirova, I, Dian, C, Leonard, G.A, Mcsweeney, S, Brzezinski, P. | | Deposit date: | 2007-04-04 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In Vivo and in Vitro Investigation of Transcriptional Regulation by Dntr.
J.Mol.Biol., 372, 2007
|
|
5XV4
 
 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
4PQI
 
 | | Crystal structure of glutathione transferase lambda3 from Populus trichocarpa | | Descriptor: | CALCIUM ION, GLUTATHIONE, In2-1 family protein, ... | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
4A2N
 
 | | Crystal Structure of Ma-ICMT | | Descriptor: | CARDIOLIPIN, ISOPRENYLCYSTEINE CARBOXYL METHYLTRANSFERASE, PALMITIC ACID, ... | | Authors: | Yang, J, Kulkarni, K, Manolaridis, I, Zhang, Z, Dodd, R.B, Mas-Droux, C, Barford, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Isoprenylcysteine Carboxyl Methylation from the Crystal Structure of the Integral Membrane Methyltransferase Icmt.
Mol.Cell, 44, 2011
|
|
4AUR
 
 | |
4TPS
 
 | | Sporulation Inhibitor of DNA Replication, SirA, in complex with Domain I of DnaA | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, Chromosomal replication initiator protein DnaA, ... | | Authors: | Jameson, K.H, Turkenburg, J.P, Fogg, M.J, Grahl, A, Wilkinson, A.J. | | Deposit date: | 2014-06-09 | | Release date: | 2014-07-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and interactions of the Bacillus subtilis sporulation inhibitor of DNA replication, SirA, with domain I of DnaA.
Mol.Microbiol., 93, 2014
|
|
4RVR
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex WITH GSK2801 | | Descriptor: | 1,2-ETHANEDIOL, 1-{1-[2-(methylsulfonyl)phenyl]-7-propoxyindolizin-3-yl}ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-11-27 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
6F2G
 
 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | Nanobody 74, Putative amino acid/polyamine transport protein, ZINC ION | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
7LYI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-3 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7LYH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-1 | | Descriptor: | 3C-like proteinase, GLYCEROL, benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7ZC2
 
 | | Dipeptide and tripeptide Permease C (DtpC) | | Descriptor: | Amino acid/peptide transporter | | Authors: | Killer, M, Finocchio, G, Pardon, E, Steyaert, J, Loew, C. | | Deposit date: | 2022-03-25 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM Structure of an Atypical Proton-Coupled Peptide Transporter: Di- and Tripeptide Permease C.
Front Mol Biosci, 9, 2022
|
|
7KAT
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore ring and Sec63 FN3 double mutant, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAJ
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class with Sec62, conformation 2 (C2) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAH
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-02-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAI
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class with Sec62, conformation 1 (C1) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8AAB
 
 | | S148F mutant of blue-to-red fluorescent timer mRubyFT | | Descriptor: | mRubyFT S148F mutant of blue-to-red fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Subach, O.M, Popov, V.O, Subach, F.V. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | mRubyFT/S147I, a mutant of blue-to-red fluorescent timer
Crystallography Reports, 2022
|
|
7KAR
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec63 FN3 mutant, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAU
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore ring and Sec63 FN3 double mutant, class with Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
