2DIG
 
 | | Solusion structure of the Todor domain of human Lamin-B receptor | | Descriptor: | Lamin-B receptor | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solusion structure of the Todor domain of human Lamin-B receptor
To be Published
|
|
8C0B
 
 | | CryoEM structure of Aspergillus nidulans UTP-glucose-1-phosphate uridylyltransferase | | Descriptor: | UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Han, X, D Angelo, C, Otamendi, A, Cifuente, J.O, de Astigarraga, E, Ochoa-Lizarralde, B, Grininger, M, Routier, F.H, Guerin, M.E, Fuehring, J, Etxebeste, O, Connell, S.R. | | Deposit date: | 2022-12-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | CryoEM analysis of the essential native UDP-glucose pyrophosphorylase from Aspergillus nidulans reveals key conformations for activity regulation and function.
Mbio, 14, 2023
|
|
8C9J
 
 | | Crystal structure of human NQO1 by serial femtosecond crystallography | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Martin-Garcia, J.M, Grieco, A, Ruiz-Fresneda, M.A, Pacheco-Garcia, J.L, Pey, A, Botha, S, Ros, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme.
Lab Chip, 23, 2023
|
|
8BSG
 
 | | COMPLEX OF LEPORINE SERUM ALBUMIN WITH DICLOFENAC | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ACETATE ION, ... | | Authors: | Bujacz, A, Talaj, J, Zielinski, K. | | Deposit date: | 2022-11-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins.
Int J Mol Sci, 24, 2023
|
|
8C8P
 
 | |
5V4K
 
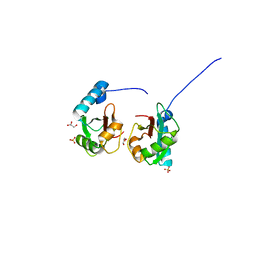 | | Crystal structure of NEDD4 LIR-fused human LC3B_2-119 | | Descriptor: | GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3B,Microtubule-associated proteins 1A/1B light chain 3B,Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Qiu, Y, Zheng, Y, Schulman, B. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Insights into links between autophagy and the ubiquitin system from the structure of LC3B bound to the LIR motif from the E3 ligase NEDD4.
Protein Sci., 26, 2017
|
|
8UWU
 
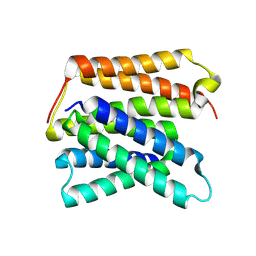 | | EmrE structure in the proton-bound state (WT/L51I heterodimer) | | Descriptor: | SMR family multidrug efflux protein EmrE | | Authors: | Li, J, Sae Her, A, Besch, A, Ramirez, B, Crames, M, Banigan, J.R, Mueller, C, Marsiglia, W.M, Zhang, Y, Traaseth, N.J. | | Deposit date: | 2023-11-08 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Dynamics underlie the drug recognition mechanism by the efflux transporter EmrE.
Nat Commun, 15, 2024
|
|
6FUJ
 
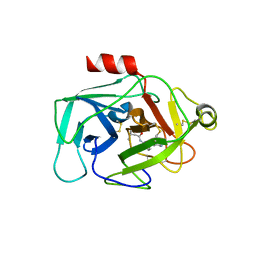 | | Complement factor D in complex with the inhibitor N-(3'-(aminomethyl)-[1,1'-biphenyl]-3-yl)-3-methylbutanamide | | Descriptor: | Complement factor D, ~{N}-[3-[3-(aminomethyl)phenyl]phenyl]-3-methyl-butanamide | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6FUI
 
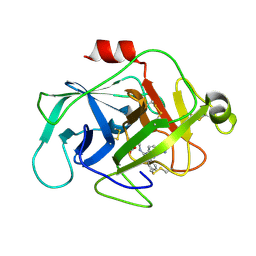 | | Complement factor D in complex with the inhibitor 3-((3-((3-(aminomethyl)phenyl)amino)-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)phenol | | Descriptor: | (1~{R},2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]cyclohexane-1-carboxylic acid, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
2OB4
 
 | | Human Ubiquitin-Conjugating Enzyme CDC34 | | Descriptor: | Ubiquitin-conjugating enzyme E2-32 kDa complementing | | Authors: | Neculai, D, Avvakumov, G.V, Xue, S, Walker, J.R, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Sicheri, F, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-18 | | Release date: | 2006-12-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
5EN3
 
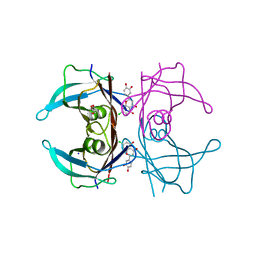 | | Crystal structure of human transthyretin in complex with luteolin-Cl at 1.25 A resolution | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-7-chloranyl-5-oxidanyl-chromen-4-one, GLYCEROL, SODIUM ION, ... | | Authors: | Begum, A, Nilsson, L, Olofsson, A, Sauer-Eriksson, A.E. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Modifications of the 7-Hydroxyl Group of the Transthyretin Ligand Luteolin Provide Mechanistic Insights into Its Binding Properties and High Plasma Specificity.
Plos One, 11, 2016
|
|
8BWT
 
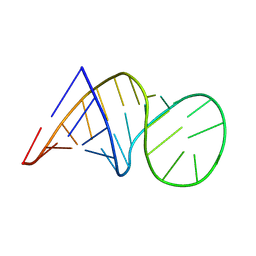 | |
6BRP
 
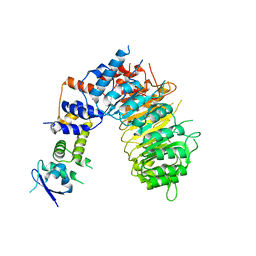 | | F-box protein form 2 | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-11-30 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
6FUG
 
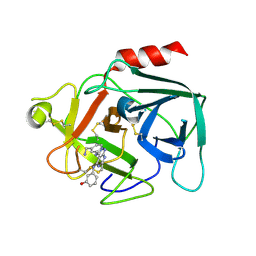 | | Complement factor D in complex with the inhibitor 3-((3-((3-(aminomethyl)phenyl)amino)-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)phenol | | Descriptor: | 3-[[3-[[3-(aminomethyl)phenyl]amino]-1~{H}-pyrazolo[3,4-d]pyrimidin-4-yl]amino]phenol, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
1HDB
 
 | | ANALYSIS OF THE CRYSTAL STRUCTURE, MOLECULAR MODELING AND INFRARED SPECTROSCOPY OF THE DISTAL BETA-HEME POCKET VALINE67(E11)-THREONINE MUTATION OF HEMOGLOBIN | | Descriptor: | HEMOGLOBIN (DEOXY) BETA-V67T, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pechik, I, Ji, X, Fronticelli, C, Gilliland, G.L. | | Deposit date: | 1995-04-14 | | Release date: | 1996-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic, molecular modeling, and biophysical characterization of the valine beta 67 (E11)-->threonine variant of hemoglobin.
Biochemistry, 35, 1996
|
|
6FUT
 
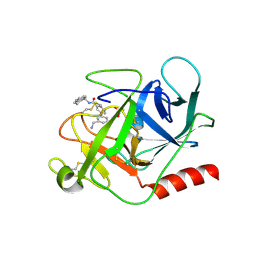 | | Complement factor D in complex with the inhibitor (S)-3'-(aminomethyl)-N-(1,2,3,4-tetrahydronaphthalen-1-yl)-[1,1'-biphenyl]-3-carboxamide | | Descriptor: | 3-[3-(aminomethyl)phenyl]-~{N}-[(1~{S})-1,2,3,4-tetrahydronaphthalen-1-yl]benzamide, Complement factor D, SUCCINIC ACID | | Authors: | Mac Sweeney, A, Vulpetti, A, Erbel, P, Lorthiois, E, Maibaum, J, Randl, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
2QGX
 
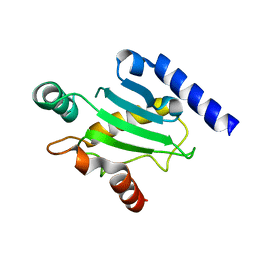 | | Ubiquitin-conjugating enzyme E2Q | | Descriptor: | Ubiquitin-conjugating enzyme E2 Q1 | | Authors: | Neculai, D, Avvakumov, G.V, Xue, S, Walker, J.R, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Sicheri, F, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
7QBR
 
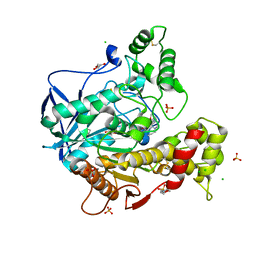 | | Human butyrylcholinesterase in complex with (Z)-N-tert-butyl-1-(8-(3-(4-(prop-2-yn-1-yl)piperazin-1-yl)propoxy)quinolin-2-yl)methanimine oxide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Denic, M, Chioua, M, Knez, D, Gobec, S, Nachon, F, Marco-Contelles, J.L, Brazzolotto, X. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | 8-Hydroxyquinolylnitrones as multifunctional ligands for the therapy of neurodegenerative diseases.
Acta Pharm Sin B, 13, 2023
|
|
7QBQ
 
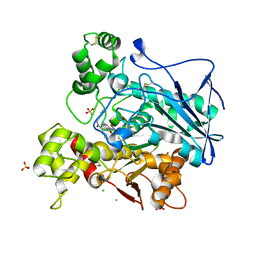 | | Human butyrylcholinesterase in complex with (Z)-N-benzyl-1-(8-hydroxyquinolin-2-yl)methanimine oxide | | Descriptor: | 1-(8-oxidanylquinolin-2-yl)-N-(phenylmethyl)methanimine oxide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Denic, M, Chioua, M, Knez, D, Gobec, S, Nachon, F, Marco-Contelles, J.L, Brazzolotto, X. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | 8-Hydroxyquinolylnitrones as multifunctional ligands for the therapy of neurodegenerative diseases.
Acta Pharm Sin B, 13, 2023
|
|
8W85
 
 | | HLA-DQ2.5-gamma2 gliadin peptide in complex with DQN0385AE01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0385AE01 Fab heavy chain, DQN0385AE01 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.769 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W83
 
 | | HLA-DQ2.5-alpha1 gliadin peptide in complex with DQN0344AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0344AE02 Fab heavy chain, DQN0344AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W86
 
 | | HLA-DQ2.5-B/C hordein peptide in complex with DQN0385AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0385AE02 Fab heavy chain, DQN0385AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W84
 
 | | HLA-DQ2.5-alpha2 gliadin peptide in complex with DQN0344AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0344AE02 Fab heavy chain, DQN0344AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
1N5M
 
 | | Crystal structure of the mouse acetylcholinesterase-gallamine complex | | Descriptor: | 2,2',2"-[1,2,3-BENZENE-TRIYLTRIS(OXY)]TRIS[N,N,N-TRIETHYLETHANAMINIUM], 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Bourne, Y, Taylor, P, Radic, Z, Marchot, P. | | Deposit date: | 2002-11-06 | | Release date: | 2003-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site
EMBO J., 22, 2003
|
|
1N5R
 
 | | Crystal structure of the mouse acetylcholinesterase-propidium complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,8-DIAMINO-5[3-(DIETHYLMETHYLAMMONIO)PROPYL]-6-PHENYLPHENANTHRIDINIUM, ACETIC ACID, ... | | Authors: | Bourne, Y, Taylor, P, Radic, Z, Marchot, P. | | Deposit date: | 2002-11-07 | | Release date: | 2003-02-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site
EMBO J., 22, 2003
|
|
