3D3P
 
 | |
3S0I
 
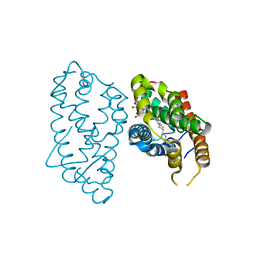 | | Crystal Structure of D48V mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo galactosylceramide containing nervonoyl acyl chain | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Popov, A.N, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
6BKE
 
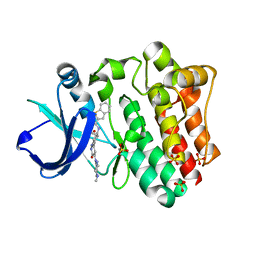 | | BTK complex with compound 10 | | Descriptor: | N-[2-(2-hydroxyethyl)-3-{5-[(5-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl)amino]-6-oxo-1,6-dihydropyridazin-3-yl}phenyl]-1-benzothiophene-2-carboxamide, SULFATE ION, Tyrosine-protein kinase BTK | | Authors: | Kiefer, J.R, Eigenbrot, C, Yu, C.L, Wang, G.X. | | Deposit date: | 2017-11-08 | | Release date: | 2018-11-07 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
9F15
 
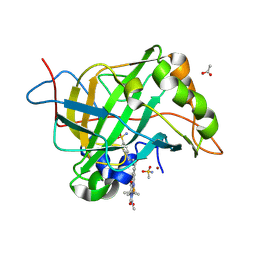 | | Carbonic anhydrase II variant with bound iron complex | | Descriptor: | 4,4,7,10,10-pentamethyl-3,6,8,11-tetrakis(oxidanylidene)-N-[2-(4-sulfamoylphenyl)ethyl]-2,5,7,9,12-pentazabicyclo[11.4.0]heptadeca-1(13),14,16-triene-15-carboxamide containing iron, ACETATE ION, Carbonic anhydrase 2, ... | | Authors: | Zhang, K, Jakob, R.P, Ward, T.R. | | Deposit date: | 2024-04-18 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Directed Evolution of an Artificial Hydroxylase Based on a Thermostable Human Carbonic Anhydrase Protein
Acs Catalysis, 14, 2024
|
|
9F2E
 
 | | Carbonic anhydrase II variant with bound iron complex in space group C2 (ArPase) | | Descriptor: | 1,2-ETHANEDIOL, 4,4,7,10,10-pentamethyl-3,6,8,11-tetrakis(oxidanylidene)-N-[2-(4-sulfamoylphenyl)ethyl]-2,5,7,9,12-pentazabicyclo[11.4.0]heptadeca-1(13),14,16-triene-15-carboxamide containing iron, Carbonic anhydrase 2, ... | | Authors: | Jakob, R.P, Ward, T.R. | | Deposit date: | 2024-04-23 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Directed Evolution of an Artificial Hydroxylase Based on a Thermostable Human Carbonic Anhydrase Protein
Acs Catalysis, 14, 2024
|
|
6C2M
 
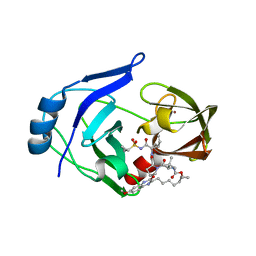 | | Crystal structure of HCV NS3/4A protease variant Y56H in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, SULFATE ION, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Clinical signature variant of HCV NS3/4A protease uses a novel mechanism to confer resistance
To Be Published
|
|
9ESC
 
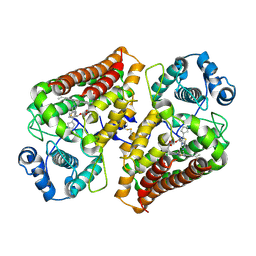 | | Holo IDO with a bound inhibitor | | Descriptor: | (~{S})-[(7~{a}~{R})-2-cyclopentyl-5,6,7,7~{a}-tetrahydroimidazo[5,1-b][1,3]thiazol-3-yl]-cyclohexyl-methanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wicki, M, Mac Sweeney, A. | | Deposit date: | 2024-03-26 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SAR and cellular potency optimization of novel heme-binding IDO1 inhibitors
To Be Published
|
|
6BKH
 
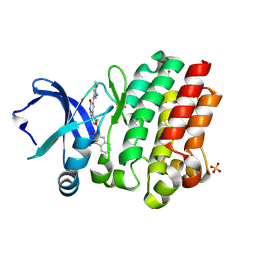 | | BTK complex with compound 11 | | Descriptor: | N-[2-(hydroxymethyl)-3-{5-[(5-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl)amino]-6-oxo-1,6-dihydropyridazin-3-yl}phenyl]-1-benzothiophene-2-carboxamide, SULFATE ION, Tyrosine-protein kinase BTK | | Authors: | Kiefer, J.R, Eigenbrot, C, Yu, C.L. | | Deposit date: | 2017-11-08 | | Release date: | 2018-11-07 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
7KK3
 
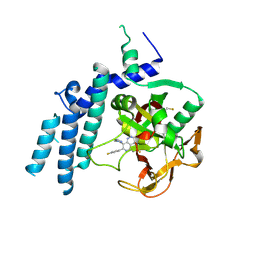 | | Structure of the catalytic domain of PARP1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
9FCO
 
 | | Structure of E. coli 30S-IF1-IF3-mRNA-Kasugamycin complex | | Descriptor: | (1S,2R,3S,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL 2-AMINO-4-{[CARBOXY(IMINO)METHYL]AMINO}-2,3,4,6-TETRADEOXY-ALPHA-D-ARABINO-HEXOPYRANOSIDE, 16S rRNA, MAGNESIUM ION, ... | | Authors: | Safdari, H.A, Wilson, D.N. | | Deposit date: | 2024-05-15 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | The translation inhibitors kasugamycin, edeine and GE81112 target distinct steps during 30S initiation complex formation.
Nat Commun, 16, 2025
|
|
7XMB
 
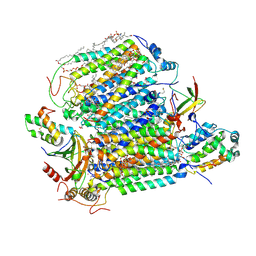 | | Crystal structure of Bovine heart cytochrome c oxidase, the structure complexed with an allosteric inhibitor T113 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Shintani, Y, Takashima, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
4LXK
 
 | | Crystal Structure of Human Beta Secretase in Complex with compound 11d | | Descriptor: | (1R,3S,4S,5R)-3-(4-amino-3-fluoro-5-{[(2R)-1,1,1-trifluoro-3-methoxypropan-2-yl]oxy}benzyl)-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1-oxide, Beta-secretase 1, GLYCEROL | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of cyclic sulfoxide hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6ZKE
 
 | | Complex I during turnover, open2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
7XMA
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, apo structure with DMSO | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
6ZKS
 
 | | Deactive complex I, open1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
3HBP
 
 | | The crystal structure of C185S mutant of recombinant sulfite oxidase with bound substrate, sulfite, at the active site | | Descriptor: | HYDROXY(DIOXO)MOLYBDENUM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, SULFITE ION, ... | | Authors: | Qiu, J.A. | | Deposit date: | 2009-05-04 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structures of the C185S and C185A mutants of sulfite oxidase reveal rearrangement of the active site.
Biochemistry, 49, 2010
|
|
5U6C
 
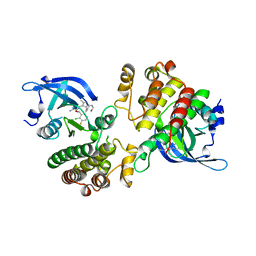 | | Crystal structure of the Mer kinase domain in complex with a macrocyclic inhibitor | | Descriptor: | (10R)-7-amino-11-chloro-12-fluoro-1-(2-hydroxyethyl)-3,10,16-trimethyl-16,17-dihydro-1H-8,4-(azeno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecin-15(10H)-one, Tyrosine-protein kinase Mer | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Axl kinase domain in complex with a macrocyclic inhibitor offers first structural insights into an active TAM receptor kinase.
J. Biol. Chem., 292, 2017
|
|
8CJ4
 
 | | Crystal structure of ClpP from Staphylococcus epidermidis, tetradecamer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Alves Franca, B, Rohde, H, Betzel, C. | | Deposit date: | 2023-02-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insights into the dynamic modulation of bacterial ClpP function and oligomerization by peptidomimetic boronate compounds.
Sci Rep, 14, 2024
|
|
5Q1E
 
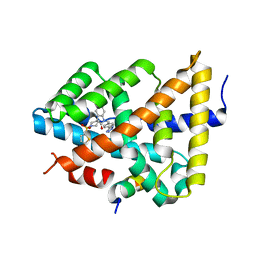 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 5-bromo-1-{[4-(1H-tetrazol-5-yl)phenyl]methyl}-1'-(thiophene-2-sulfonyl)spiro[indole-3,4'-piperidin]-2(1H)-one, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
3GOK
 
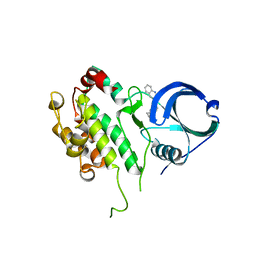 | | Binding site mapping of protein ligands | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 2 | | Authors: | Scheich, C. | | Deposit date: | 2009-03-19 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Binding site mapping of protein ligands
To be published
|
|
6ZKQ
 
 | | Native complex I, open2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
7KFP
 
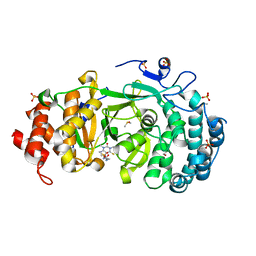 | | Structure of human PARG complexed with PARG-119 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{[2-(1,3-dimethyl-2-oxo-6-sulfanylidene-1,2,3,6-tetrahydro-7H-purin-7-yl)ethyl]carbamoyl}methanesulfonamide, ... | | Authors: | Brosey, C.A, Bommagani, S, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6ZKJ
 
 | | Complex I with NADH, open3 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
3GPE
 
 | | Crystal Structure Analysis of PKC (alpha)-C2 domain complexed with Ca2+ and PtdIns(4,5)P2 | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Protein kinase C alpha type, ... | | Authors: | Ferrer-Orta, C, Querol-Audi, J, Fita, I, Verdaguer, N. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7KG0
 
 | | Structure of human PARG complexed with PARG-131 | | Descriptor: | 1,2-ETHANEDIOL, 5-({4-[(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)methyl]phenyl}methyl)pyrimidine-2,4,6(1H,3H,5H)-trione, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Arvai, A, Bommagani, S, Brosey, C.A, Jones, D.E, Warden, L.S, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-15 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
