8OUQ
 
 | | Clr-11 from Rattus norvegicus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D11 | | Authors: | Skalova, T, Blaha, J, Kalouskova, B, Skorepa, O, Vanek, O, Dohnalek, J. | | Deposit date: | 2023-04-24 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Clr-11 from Rattus norvegicus
To Be Published
|
|
6O63
 
 | |
4WFC
 
 | | Structure of the Rrp6-Rrp47 interaction | | Descriptor: | Exosome complex exonuclease RRP6, Exosome complex protein LRP1, SULFATE ION | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
5I4V
 
 | | Discovery of novel, orally efficacious Liver X Receptor (LXR) beta agonists | | Descriptor: | Oxysterols receptor LXR-beta,Nuclear receptor coactivator 2, Retinoic acid receptor RXR-beta,Nuclear receptor coactivator 2, {2-[(2R)-4-[4-(hydroxymethyl)-3-(methylsulfonyl)phenyl]-2-(propan-2-yl)piperazin-1-yl]-4-(trifluoromethyl)pyrimidin-5-yl}methanol | | Authors: | Chen, G, McKeever, B.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of a Novel, Orally Efficacious Liver X Receptor (LXR) beta Agonist.
J.Med.Chem., 59, 2016
|
|
7N6P
 
 | | Crystal structure of the anti-EBOV and SUDV monoclonal antibody 1C3 Fab | | Descriptor: | 1C3 Fab heavy chain, 1C3 Fab light chain | | Authors: | Milligan, J.C, Yu, X, Buck, T, Saphire, E.O. | | Deposit date: | 2021-06-08 | | Release date: | 2022-04-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Asymmetric and non-stoichiometric glycoprotein recognition by two distinct antibodies results in broad protection against ebolaviruses.
Cell, 185, 2022
|
|
8SWF
 
 | | Cryo-EM structure of NLRP3 open octamer | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
5IF4
 
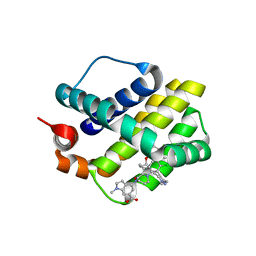 | | Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) inhibitors using Structure-Based Design | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Discovery and biological characterization of potent myeloid cell leukemia-1 inhibitors.
FEBS Lett., 591, 2017
|
|
5LAL
 
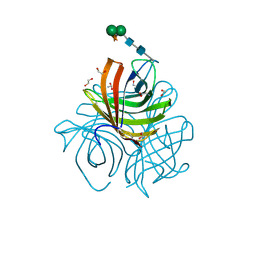 | | Structure of Arabidopsis dirigent protein AtDIR6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dirigent protein 6, ... | | Authors: | Gasper, R, Kolesinski, P, Terlecka, B, Effenberger, I, Schaller, A, Hofmann, E. | | Deposit date: | 2016-06-14 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dirigent Protein Mode of Action Revealed by the Crystal Structure of AtDIR6.
Plant Physiol., 172, 2016
|
|
6OFZ
 
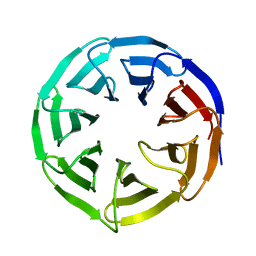 | | Crystal structure of human WDR5 | | Descriptor: | WD repeat-containing protein 5 | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
4WSP
 
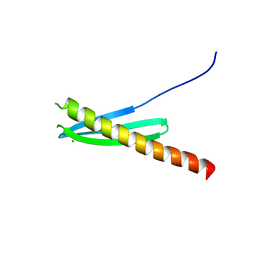 | | Racemic crystal structure of Rv1738 from Mycobacterium tuberculosis (Form-I) | | Descriptor: | CHLORIDE ION, protein DL-Rv1738 | | Authors: | Bunker, R.D, Mandal, K, Kent, S.B.H, Baker, E.N. | | Deposit date: | 2014-10-28 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A functional role of Rv1738 in Mycobacterium tuberculosis persistence suggested by racemic protein crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6OIP
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 34 | | Descriptor: | 2-fluoro-3-methyl-N'-[(naphthalen-2-yl)sulfonyl]benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Benzoylsulfonohydrazides as Potent Inhibitors of the Histone Acetyltransferase KAT6A.
J.Med.Chem., 62, 2019
|
|
7AJP
 
 | | Crystal Structure of Human Adenovirus 56 Fiber Knob | | Descriptor: | 1,2-ETHANEDIOL, Fiber, GLYCEROL, ... | | Authors: | Strebl, M, Mindler, K, Stehle, T. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.382018 Å) | | Cite: | Human species D adenovirus hexon capsid protein mediates cell entry through a direct interaction with CD46.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ALI
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup P2(1)). | | Descriptor: | 3C-like proteinase | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | Deposit date: | 2020-10-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
6Y2B
 
 | | Crystal structure of HLA-B2709 complexed with the nona-peptide mQ | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Lymphocyte antigen HLA-B27, ... | | Authors: | Loll, B, Rueckert, C, Ziegler, B.-U, Ziegler, A. | | Deposit date: | 2020-02-15 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A CENTRAL PEPTIDE RESIDUE CAN CONTROL
MHC POLYMORPHISM-DEPENDENT ANTIGEN PRESENTATION
to be published
|
|
4X5R
 
 | | Crystal structure of FimH in complex with a squaryl-phenyl alpha-D-mannopyranoside derivative | | Descriptor: | 2-chloro-4-{[2-(4-methylpiperazin-1-yl)-3,4-dioxocyclobut-1-en-1-yl]amino}phenyl alpha-D-mannopyranoside, Protein FimH, SULFATE ION | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
7AXL
 
 | | Crystal structure of the hPXR-LBD in complex with estradiol and heptachlor endo-epoxide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ESTRADIOL, Nuclear receptor subfamily 1 group I member 2, ... | | Authors: | Delfosse, V, Granell, M, Blanc, P, Bourguet, W. | | Deposit date: | 2020-11-09 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic insights into the synergistic activation of the RXR-PXR heterodimer by endocrine disruptor mixtures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ENG
 
 | | Crystal structure of the 43K ATPase domain of Escherichia coli gyrase B in complex with an aminocoumarin | | Descriptor: | CHLORIDE ION, Coumermycin A1, DNA gyrase subunit B, ... | | Authors: | Vanden Broeck, A, McEwen, A.G, Lamour, V. | | Deposit date: | 2017-10-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for DNA Gyrase Interaction with Coumermycin A1.
J.Med.Chem., 62, 2019
|
|
4WIG
 
 | | Crystal structure of E47D mutant cytidine deaminase from Mycobacterium tuberculosis (MtCDA E47D) | | Descriptor: | Cytidine deaminase, ZINC ION | | Authors: | Trivella, D.B.B, Sanchez-Quitian, A.Z, Basso, L.A, Santos, D.S. | | Deposit date: | 2014-09-25 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | Functional and structural evidence for the catalytic role played by glutamate-47 residue in the mode of action of Mycobacterium tuberculosis cytidine deaminase
Rsc Adv, 5, 2015
|
|
7TE1
 
 | |
8PA2
 
 | |
7N5Q
 
 | |
6YES
 
 | |
4WRK
 
 | |
8PA1
 
 | |
1BC8
 
 | | STRUCTURES OF SAP-1 BOUND TO DNA SEQUENCES FROM THE E74 AND C-FOS PROMOTERS PROVIDE INSIGHTS INTO HOW ETS PROTEINS DISCRIMINATE BETWEEN RELATED DNA TARGETS | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*TP*CP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*GP*GP*AP*AP*GP*T)-3'), PROTEIN (SAP-1 ETS DOMAIN), ... | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 1998-05-05 | | Release date: | 1998-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins.
Mol.Cell, 2, 1998
|
|
