8S1W
 
 | |
1R6C
 
 | | High resolution structure of ClpN | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the
ClpA chaperone
J.Struct.Biol., 146, 2004
|
|
1RAX
 
 | |
8SCC
 
 | | Crystal Structure of L-galactose 1-dehydrogenase de Myrciaria dubia | | Descriptor: | L-galactose dehydrogenase | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-05 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
1R6K
 
 | | HPV11 E2 TAD crystal structure | | Descriptor: | HPV11 REGULATORY PROTEIN E2 | | Authors: | Wang, Y, Coulombe, R. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the E2 Transactivation Domain of Human Papillomavirus Type 11 Bound to a Protein Interaction Inhibitor
J.Biol.Chem., 279, 2004
|
|
8SG0
 
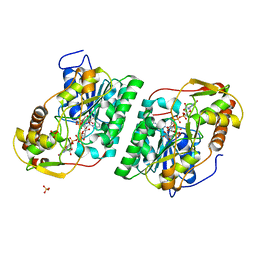 | | Crystal Structure of GDP-manose 3,5 epimerase de Myrciaria dubia in complex with substrate, product and NAD | | Descriptor: | GDP-mannose 3,5-epimerase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, GUANOSINE-5'-DIPHOSPHATE-BETA-L-GALACTOSE, ... | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
1R6Y
 
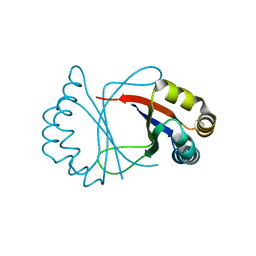 | |
8SLN
 
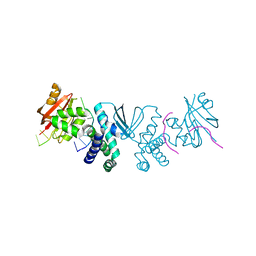 | |
1R7L
 
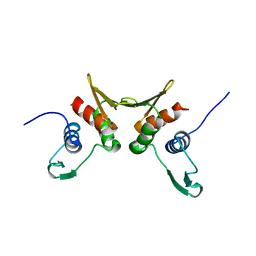 | | 2.0 A Crystal Structure of a Phage Protein from Bacillus cereus ATCC 14579 | | Descriptor: | Phage protein | | Authors: | Zhang, R, Joachimiak, G, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of phage protein BC1872 from Bacillus cereus, a singleton with new fold
Proteins, 64, 2006
|
|
8S8W
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA-RNA (Cap0-RNA) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-07 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
1RD5
 
 | | Crystal structure of Tryptophan synthase alpha chain homolog BX1: a member of the chemical plant defense system | | Descriptor: | MALONIC ACID, Tryptophan synthase alpha chain, chloroplast | | Authors: | Kulik, V, Hartmann, E, Weyand, M, Frey, M, Gierl, A, Niks, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2003-11-05 | | Release date: | 2004-12-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes.
J.Mol.Biol., 352, 2005
|
|
8SNF
 
 | | Crystal structure of metformin hydrolase (MfmAB) from Pseudomonas mendocina sp. MET-2 with Ni2+2 bound | | Descriptor: | NICKEL (II) ION, metformin hydrolase subunit A, metformin hydrolase subunit B | | Authors: | Tassoulas, L.J, Rankin, J.A, Elias, M.H, Wackett, L.P. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dinickel enzyme evolved to metabolize the pharmaceutical metformin and its implications for wastewater and human microbiomes.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1R8P
 
 | | HPV-16 E2C solution structure | | Descriptor: | Regulatory protein E2 | | Authors: | Nadra, A.D, Eliseo, T, Cicero, D.O. | | Deposit date: | 2003-10-28 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HPV-16 E2 DNA binding domain, a transcriptional regulator with a dimeric beta-barrel fold
J.Biomol.NMR, 30, 2004
|
|
8SXG
 
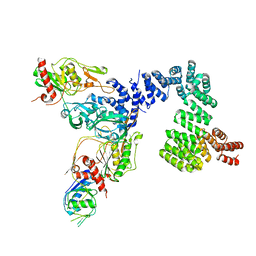 | |
1RAH
 
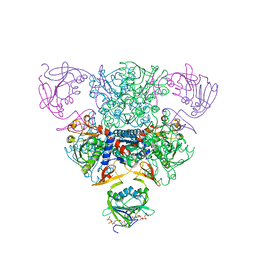 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1RBU
 
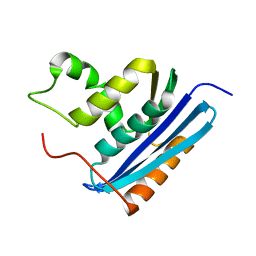 | | STRUCTURAL STUDY OF MUTANTS OF ESCHERICHIA COLI RIBONUCLEASE HI WITH ENHANCED THERMOSTABILITY | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Kanaya, S, Morikawa, K, Nakamura, H. | | Deposit date: | 1993-02-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability.
Protein Eng., 6, 1993
|
|
1REJ
 
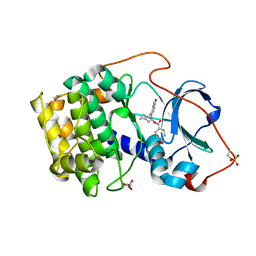 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 1 | | Descriptor: | 3-[(4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-HYDROXYBENZOATE, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
8SXH
 
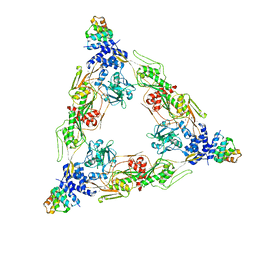 | |
1RFL
 
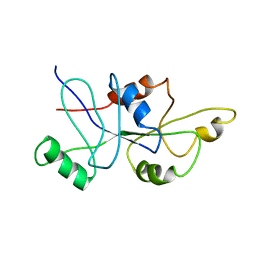 | | NMR data driven structural model of G-domain of MnmE protein | | Descriptor: | Probable tRNA modification GTPase trmE | | Authors: | Monleon, D, Esteve, V, Martinez-Vicente, M, Yim, L, Armengod, M.E, Celda, B. | | Deposit date: | 2003-11-10 | | Release date: | 2003-12-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the GTPase domain of Escherichia coli MnmE protein.
Proteins, 66, 2007
|
|
6WJ5
 
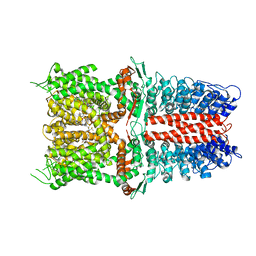 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
1RFY
 
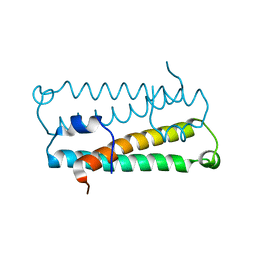 | | Crystal Structure of Quorum-Sensing Antiactivator TraM | | Descriptor: | Transcriptional repressor traM | | Authors: | Chen, G, Malenkos, J.W, Cha, M.R, Fuqua, C, Chen, L. | | Deposit date: | 2003-11-10 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR
Mol.Microbiol., 52, 2004
|
|
1RL1
 
 | | Solution structure of human Sgt1 CS domain | | Descriptor: | Suppressor of G2 allele of SKP1 homolog | | Authors: | Lee, Y.-T, Jacob, J, Michowski, W, Nowotny, M, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2003-11-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human Sgt1 Binds HSP90 through the CHORD-Sgt1 Domain and Not the Tetratricopeptide Repeat Domain
J.Biol.Chem., 279, 2004
|
|
1RM5
 
 | | Crystal structure of mutant S188A of photosynthetic glyceraldehyde-3-phosphate dehydrogenase A4 isoform, complexed with NADP | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Sparla, F, Fermani, S, Falini, G, Ripamonti, A, Sabatino, P, Pupillo, P, Trost, P. | | Deposit date: | 2003-11-27 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Coenzyme Site-directed Mutants of Photosynthetic A(4)-GAPDH Show Selectively Reduced NADPH-dependent Catalysis, Similar to Regulatory AB-GAPDH Inhibited by Oxidized Thioredoxin
J.Mol.Biol., 340, 2004
|
|
8S8X
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Toyocamycin and m7GpppA-RNA (Cap0-RNA) | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-07 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
8S9L
 
 | | Structure of monomeric FAM111A SPD V347D Mutant | | Descriptor: | SULFATE ION, Serine protease FAM111A | | Authors: | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
