6T8U
 
 | | Complement factor B in complex with 5-Bromo-3-chloro-N-(4,5-dihydro-1H-imidazol-2-yl)-7-methyl-1H-indol-4-amine | | Descriptor: | 5-bromanyl-3-chloranyl-~{N}-(1~{H}-imidazol-2-yl)-7-methyl-1~{H}-indol-4-amine, Complement factor B, SULFATE ION | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6T8V
 
 | | Complement factor B in complex with (S)-5,7-Dimethyl-4-((2-phenylpiperidin-1-yl)methyl)-1H-indole | | Descriptor: | 4-[(2~{S})-1-[(5,7-dimethyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6L5Y
 
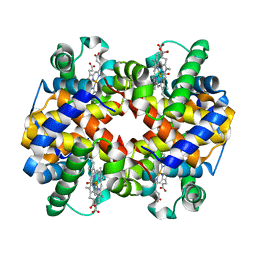 | | Carbonmonoxy human hemoglobin A in the R2 quaternary structure at 140 K: Light (2 min) | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L5W
 
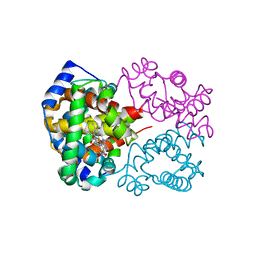 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 140 K: Light (2 min) | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L5X
 
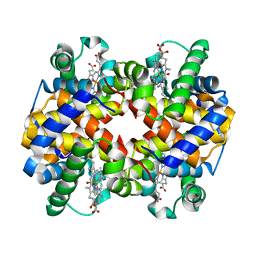 | | Carbonmonoxy human hemoglobin A in the R2 quaternary structure at 95 K: Light (2 min) | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L5V
 
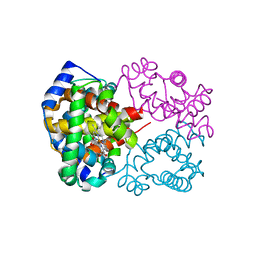 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 95 K: Light (2 min) | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6URQ
 
 | | Complex structure of human poly-ADP-ribosyltransferase TNKS1 ARC2-ARC3 and P antigen family member 4 (PAGE4) | | Descriptor: | GLYCEROL, P antigen family member 4, Poly [ADP-ribose] polymerase tankyrase-1, ... | | Authors: | Zheng, Y, Koirala, S, Miller, D, Potts, P.R. | | Deposit date: | 2019-10-24 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tissue-Specific Regulation of the Wnt/ beta-Catenin Pathway by PAGE4 Inhibition of Tankyrase.
Cell Rep, 32, 2020
|
|
6URT
 
 | |
6T89
 
 | | Thrombin in complex with (S)-N-(tert-butyl)-4-(3-(3-carbamimidoylphenyl)-2-((2',4'-dimethoxy-[1,1'-biphenyl])-3-sulfonamido)propanoyl)piperazine-1-carboxamide (MI-498) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(2~{S})-3-(3-carbamimidoylphenyl)-2-[[3-(4-methoxy-2-oxidanyl-phenyl)phenyl]sulfonylamino]propanoyl]-~{N}-methyl-piperazine-1-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Ngaha, S.A, Sandner, A, Huber, S, Heine, A, Steinmetzer, T, Pilgram, O. | | Deposit date: | 2019-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improving the selectivity of 3-amidinophenylalanine-derived matriptase inhibitors
Eur.J.Med.Chem., 2022
|
|
6T8A
 
 | | Thrombin in complex with diphenyl ((4-carbamimidoylphenyl)((S)-1-((R)-3-cyclohexyl 2-((phenylmethyl)sulfonamido)propanoyl)pyrrolidine-2-carboxamido)methyl)phosphonate (MI-492) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Hirudin variant-2, ... | | Authors: | Ngaha, S.A, Sandner, A, Huber, S, Heine, A, Steinmetzer, T, Pilgram, O. | | Deposit date: | 2019-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Thrombin in complex with diphenyl ((4-carbamimidoylphenyl)((S)-1-((R)-3-cyclohexyl-2-((phenylmethyl)sulfonamido)propanoyl)pyrrolidine-2-carboxamido)methyl)phosphonate (MI-492)
to be published
|
|
6T8N
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K3007 | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type I, DIMETHYL SULFOXIDE, ... | | Authors: | Adamson, R.J, Williams, E.P, Bonomo, S, Rankin, S, Bacos, D, Rae, A, Cramp, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-24 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K3007
To Be Published
|
|
6URA
 
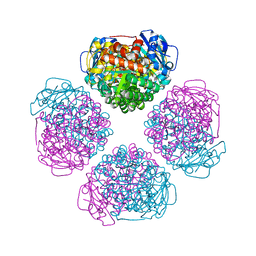 | | Crystal structure of RUBISCO from Promineofilum breve | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain | | Authors: | Pereira, J.H, Banda, D.M, Liu, A.K, Shih, P.M, Adams, P.D. | | Deposit date: | 2019-10-23 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel bacterial clade reveals origin of form I Rubisco.
Nat.Plants, 6, 2020
|
|
6URG
 
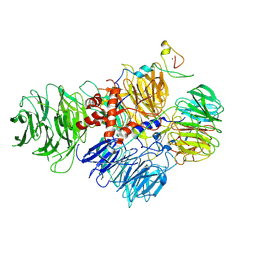 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-CPSF100 PIM complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 4, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
6URC
 
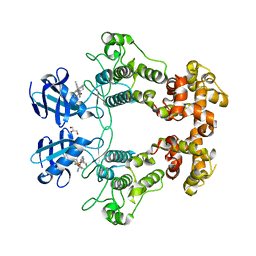 | | Crystal structure of IRE1a in complex with compound 18 | | Descriptor: | 2-chloro-N-(6-methyl-5-{[3-(2-{[(3S)-piperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}naphthalen-1-yl)benzene-1-sulfonamide, GLYCEROL, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H.H, Wang, W. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Disruption of IRE1 alpha through its kinase domain attenuates multiple myeloma.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6URO
 
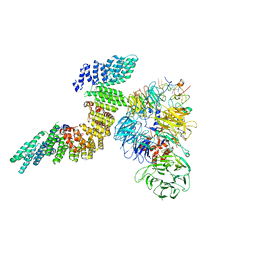 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-PAS RNA-CstF77 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, Cleavage stimulation factor subunit 3, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
6T7P
 
 | | human plasmakallikrein protease domain in complex with active site directed inhibitor | | Descriptor: | (2~{S},4~{R})-1-[[(3~{S})-3-azanyl-2,3-dihydro-1-benzofuran-6-yl]carbonyl]-~{N}-(3-chlorophenyl)-4-phenyl-pyrrolidine-2-carboxamide, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Renatus, M. | | Deposit date: | 2019-10-22 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.416 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6T7H
 
 | | Crystal structure of Thrombin in complex with macrocycle N14-PR4-A | | Descriptor: | (14S,17R)-14-(3-carbamimidamidopropyl)-3-(furan-2-ylmethyl)-5,12,15-tris(oxidanylidene)-19-thia-3,6,13,16-tetrazatricyclo[19.4.0.0^{6,10}]pentacosa-1(25),7,9,21,23-pentaene-17-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Angelini, A, Kumar, M.G, Heinis, C, Cendron, L. | | Deposit date: | 2019-10-22 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Macrocycle synthesis strategy based on step-wise "adding and reacting" three components enables screening of large combinatorial libraries.
Chem Sci, 11, 2020
|
|
6UQO
 
 | | ClpA/ClpP Engaged State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp endopeptidase proteolytic subunit ClpP, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Southworth, D.R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-04-22 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UQF
 
 | | Human HCN1 channel in a hyperpolarized conformation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, MERCURY (II) ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lee, C.-H, MacKinnon, R. | | Deposit date: | 2019-10-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Voltage Sensor Movements during Hyperpolarization in the HCN Channel.
Cell, 179, 2019
|
|
6UQG
 
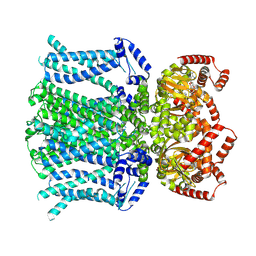 | | Human HCN1 channel Y289D mutant | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lee, C.-H, MacKinnon, R. | | Deposit date: | 2019-10-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Voltage Sensor Movements during Hyperpolarization in the HCN Channel.
Cell, 179, 2019
|
|
6UQE
 
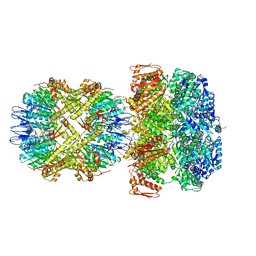 | | ClpA/ClpP Disengaged State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Lopez, K.L, Rizo, A.R, Southworth, D.R. | | Deposit date: | 2019-10-18 | | Release date: | 2020-04-22 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UPV
 
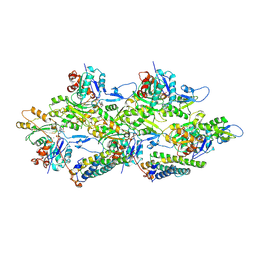 | | Alpha-E-catenin ABD-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mei, L, Alushin, G.M. | | Deposit date: | 2019-10-18 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular mechanism for direct actin force-sensing by alpha-catenin.
Elife, 9, 2020
|
|
6UPW
 
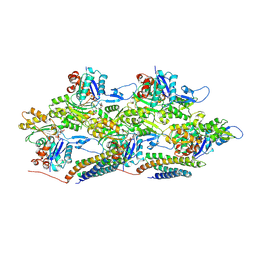 | | Metavinculin ABD-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mei, L, Alushin, G.M. | | Deposit date: | 2019-10-18 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for direct actin force-sensing by alpha-catenin.
Elife, 9, 2020
|
|
6T6F
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 8 (CS275) | | Descriptor: | 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3-(trifluoromethyl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D | | Authors: | Sorrell, F, Kraemer, A, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | CAMK1D bound to CS275
To Be Published
|
|
6T6D
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2149 | | Descriptor: | 2-methoxy-4-[4-methyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]benzamide, Activin receptor type I, SULFATE ION | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Targeting ALK2: An Open Science Approach to Developing Therapeutics for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
