6QLA
 
 | | CRYSTAL STRUCTURE OF THE PMGL2 ESTERASE (point mutant 1) FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Boyko, K.M, Garsia, D, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of PMGL2 esterase from the hormone-sensitive lipase family with GCSAG motif around the catalytic serine.
Plos One, 15, 2020
|
|
4X1L
 
 | |
6O5X
 
 | | Crystal Structure of multi-drug resistant HIV-1 protease PR-S17 with substrate analog CA-p2 | | Descriptor: | CHLORIDE ION, HIV-1 protease PR-S17, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, ... | | Authors: | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2019-03-04 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Highly Drug-Resistant HIV-1 Protease Mutant PRS17 Shows Enhanced Binding to Substrate Analogues.
Acs Omega, 4, 2019
|
|
8D4I
 
 | | Structure of Y430F D-ornithine/D-lysine decarboxylase complex with putrescine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Phillips, R.S, Nguyen Hoang, K.N. | | Deposit date: | 2022-06-02 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The Y430F mutant of Salmonella d-ornithine/d-lysine decarboxylase has altered stereospecificity and a putrescine allosteric activation site.
Arch.Biochem.Biophys., 731, 2022
|
|
8ET9
 
 | |
6OB0
 
 | | Compound 2 bound structure of WT Lipoprotein Lipase in Complex with GPIHBP1 Mutant N78D N82D produced in HEK293-F cells | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Arora, R, Horton, P.A, Benson, T.E, Romanowski, M.J. | | Deposit date: | 2019-03-19 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure of lipoprotein lipase in complex with GPIHBP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OBH
 
 | | Structure of HIV-1 CA 1/2-hexamer | | Descriptor: | CA, SODIUM ION | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2019-03-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
8D88
 
 | |
4X1M
 
 | |
8K13
 
 | | SID1 transmembrane family member 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8D5R
 
 | | Structure of Y430F D-ornithine/D-lysine decarboxylase complex with D-ornithine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ACETATE ION, ... | | Authors: | Phillips, R.S, Nguyen Hoang, K.N. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The Y430F mutant of Salmonella d-ornithine/d-lysine decarboxylase has altered stereospecificity and a putrescine allosteric activation site.
Arch.Biochem.Biophys., 731, 2022
|
|
8K1D
 
 | | SID1 transmembrane family member 1 | | Descriptor: | SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8D2Y
 
 | | Y430F mutant of D-ornithine/D-lysine decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Phillips, R.S, Nguyen Hoang, K.N. | | Deposit date: | 2022-05-31 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | The Y430F mutant of Salmonella d-ornithine/d-lysine decarboxylase has altered stereospecificity and a putrescine allosteric activation site.
Arch.Biochem.Biophys., 731, 2022
|
|
4WXX
 
 | | The crystal structure of human DNMT1(351-1600) | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Zhang, Z.M, Song, J. | | Deposit date: | 2014-11-14 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.622 Å) | | Cite: | Crystal Structure of Human DNA Methyltransferase 1.
J.Mol.Biol., 427, 2015
|
|
4X9J
 
 | | EGR-1 with Doubly Methylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*(5CM)P*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*(5CM)P*GP*C)-3'), Early growth response protein 1, ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2014-12-11 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.412 Å) | | Cite: | Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1.
Febs Lett., 589, 2015
|
|
6OYR
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-002 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(3-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
4WY7
 
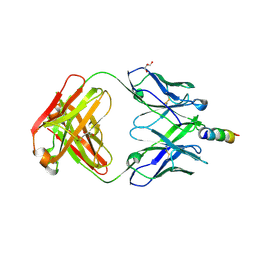 | | Crystal structure of recombinant 4E10 expressed in Escherichia coli with epitope bound | | Descriptor: | Envelope glycoprotein gp160, Fab 4E10 Heavy chain, Fab 4E10Light chain, ... | | Authors: | Rujas, E, Morante, K, Tsumoto, K, Nieva, J.L, Caaveiro, J.M.M. | | Deposit date: | 2014-11-16 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Atomic Structure of the HIV-1 gp41 Transmembrane Domain and Its Connection to the Immunogenic Membrane-proximal External Region.
J.Biol.Chem., 290, 2015
|
|
4WYM
 
 | | Structural basis of HIV-1 capsid recognition by CPSF6 | | Descriptor: | Capsid protein p24, ISOFORM 2 OF CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR SUBUNIT 6 | | Authors: | Battacharya, A, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-11-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of HIV-1 capsid recognition by PF74 and CPSF6.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
4X16
 
 | | JC Mad-1 polyomavirus VP1 in complex with GD1a oligosaccharide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein VP1, ... | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Greater Affinity of JC Polyomavirus Capsid for alpha 2,6-Linked Lactoseries Tetrasaccharide c than for Other Sialylated Glycans Is a Major Determinant of Infectivity.
J.Virol., 89, 2015
|
|
6QZ8
 
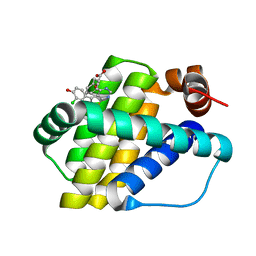 | | Structure of Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6WK1
 
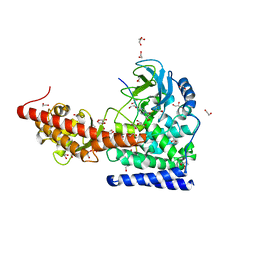 | | SETD3 in Complex with an Actin Peptide with His73 Replaced with Methionine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Characterization of SETD3 methyltransferase-mediated protein methionine methylation.
J.Biol.Chem., 295, 2020
|
|
6YAJ
 
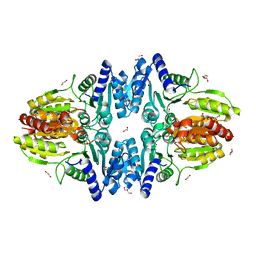 | | Split gene transketolase, inactive beta4 tetramer | | Descriptor: | 1,2-ETHANEDIOL, C-terminal chain of split transketolase from Carboxydothermus hydrogenoformans, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P. | | Deposit date: | 2020-03-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 'Split-Gene' Transketolase From the Hyper-Thermophilic Bacterium Carboxydothermus hydrogenoformans : Structure and Biochemical Characterization.
Front Microbiol, 11, 2020
|
|
4XJO
 
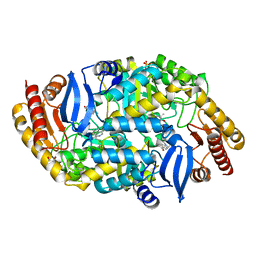 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with an inhibitor optimized from HTS lead | | Descriptor: | 1,2-ETHANEDIOL, 5-[4-(3-chlorobenzoyl)piperazin-1-yl]-1H-inden-1-one, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, ... | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2015-01-08 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization of Pyridoxal 5'-Phosphate-Dependent Transaminase Enzyme (BioA) Inhibitors that Target Biotin Biosynthesis in Mycobacterium tuberculosis.
J. Med. Chem., 60, 2017
|
|
9CDC
 
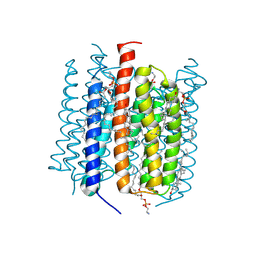 | | Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Dark State | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Kalium channelrhodopsin 1 C110A, ... | | Authors: | Morizumi, T, Kim, K, Ernst, O.P. | | Deposit date: | 2024-06-24 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural insights into light-gating of potassium-selective channelrhodopsin.
Nat Commun, 16, 2025
|
|
