2V42
 
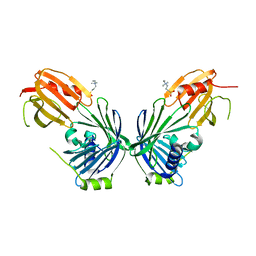 | |
5W4K
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Klebsazolicin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Metelev, M, Osterman, I.A, Ghilarov, D, Khabibullina, N.F, Yakimov, A, Shabalin, K, Utkina, I, Travin, D.Y, Komarova, E.S, Serebryakova, M, Artamonova, T, Khodorkovskii, M, Konevega, A.L, Sergiev, P.V, Severinov, K, Polikanov, Y.S. | | Deposit date: | 2017-06-12 | | Release date: | 2017-08-30 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel.
Nat. Chem. Biol., 13, 2017
|
|
7HOY
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with ASAP-0014767-001 | | Descriptor: | 3-chloro-5-[(E)-({3-[(dimethylamino)methyl]-4-hydroxyphenyl}methylidene)amino]-N-(1-hydroxy-2-methylpropan-2-yl)benzamide, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Ni, X, Marples, P.G, Godoy, A.S, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Fairhead, M, Lithgo, R.M, Lee, A, Kenton, N, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Williams, E.P, Chandran, A.V, Walsh, M.A, Fearon, D, von Delft, F. | | Deposit date: | 2024-11-07 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
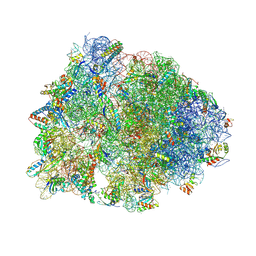
5FKX
 
 | | Structure of E.coli inducible lysine decarboxylase at active pH | | Descriptor: | LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
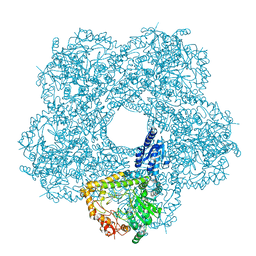
5FL2
 
 | | Revisited cryo-EM structure of Inducible lysine decarboxylase complexed with LARA domain of RavA ATPase | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-21 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
4O4K
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-2-azanylidene-5-[(4-hydroxyphenyl)methylidene]-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
6CWD
 
 | |
6CXY
 
 | |
4O43
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-3-[(2~{R})-butan-2-yl]-5-[(4-hydroxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
4O5G
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-5-[(4-aminophenyl)methylidene]-2-azanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
5ZDP
 
 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with ferulenol | | Descriptor: | 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, Alternative oxidase, mitochondrial, ... | | Authors: | Shiba, T, Inaoka, D.K, Takahashi, G, Tsuge, C, Kido, Y, Young, L, Ueda, S, Balogun, E.O, Nara, T, Honma, T, Tanaka, A, Inoue, M, Saimoto, H, Harada, S, Moore, A.L, Kita, K. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Insights into the ubiquinol/dioxygen binding and proton relay pathways of the alternative oxidase.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
7WTM
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Dis-E | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, La Venuta, G, Lau, B, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | In vitro structural maturation of an early stage pre-40S particle coupled with U3 snoRNA release and central pseudoknot formation.
Nucleic Acids Res., 50, 2022
|
|
6I2K
 
 | | Structure of EV71 complexed with its receptor SCARB2 | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Zhao, Y, Kotecha, A, Fry, E.E, Kelly, J, Wang, X, Rao, Z, Rowlands, D.J, Ren, J, Stuart, D.I. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2.
Nat Microbiol, 4, 2019
|
|
6A6K
 
 | | Crystal structure of Estrogen-related Receptor-3 (ERR-gamma) ligand binding domain with DN201000 | | Descriptor: | 3-[(~{E})-5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Cho, S.J, Song, J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Estrogen-Related Receptor-gamma Inverse Agonists To Restore the Sodium Iodide Symporter Function in Anaplastic Thyroid Cancer.
J. Med. Chem., 62, 2019
|
|
6AD9
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-9 | | Descriptor: | 12-mer peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, 3-[(1E)-1-{8-[(4-methyl-2-propyl-1H-benzimidazol-1-yl)methyl]dibenzo[b,e]oxepin-11(6H)-ylidene}ethyl]-1,2,4-oxadiazol-5(4H)-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Takahashi, Y, Suzuki, M, Yamamoto, K, Saito, J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of Dihydrodibenzooxepine Peroxisome Proliferator-Activated Receptor (PPAR) Gamma Ligands of a Novel Binding Mode as Anticancer Agents: Effective Mimicry of Chiral Structures by Olefinic E/ Z-Isomers.
J. Med. Chem., 61, 2018
|
|
5G1C
 
 | | Structure of HDAC like protein from Bordetella Alcaligenes bound the photoswitchable pyrazole Inhibitor CEW395 | | Descriptor: | (2E)-N-hydroxy-3-{4-[(E)-(1,3,5-trimethyl-1H-pyrazol-4-yl)diazenyl]phenyl}prop-2-enamide, DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Toward Photopharmacological Antimicrobial Chemotherapy Using Photoswitchable Amidohydrolase Inhibitors.
ACS Infect Dis, 3, 2017
|
|
5FKA
 
 | | Crystal structure of staphylococcal enterotoxin E in complex with a T cell receptor | | Descriptor: | STAPHYLOCOCCAL ENTEROTOXIN E, T CELL RECEPTOR ALPHA CHAIN, T CELL RECEPTOR BETA CHAIN, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2015-10-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two Common Structural Motifs for Tcr Recognition by Staphylococcal Enterotoxins.
Sci.Rep., 6, 2016
|
|
1LTA
 
 | | 2.2 ANGSTROMS CRYSTAL STRUCTURE OF E. COLI HEAT-LABILE ENTEROTOXIN (LT) WITH BOUND GALACTOSE | | Descriptor: | HEAT-LABILE ENTEROTOXIN, SUBUNIT A, SUBUNIT B, ... | | Authors: | Merritt, E.A, Sixma, T.K, Kalk, K.H, Van Zanten, B.A.M, Hol, W.G.J. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Galactose-binding site in Escherichia coli heat-labile enterotoxin (LT) and cholera toxin (CT).
Mol.Microbiol., 13, 1994
|
|
5ZMA
 
 | | Structural basis for an allosteric Eya2 phosphatase inhibitor | | Descriptor: | 3-fluoro-N'-[(E)-{5-[(pyrimidin-2-yl)sulfanyl]furan-2-yl}methylidene]benzohydrazide, Eyes absent homolog 2 | | Authors: | Anantharajan, J, Jansson, A.E, Kang, C. | | Deposit date: | 2018-04-02 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.175 Å) | | Cite: | Structural and Functional Analyses of an Allosteric EYA2 Phosphatase Inhibitor That Has On-Target Effects in Human Lung Cancer Cells.
Mol.Cancer Ther., 18, 2019
|
|
6A4S
 
 | | Crystal structure of peptidase E with ordered active site loop from Salmonella enterica | | Descriptor: | Peptidase E | | Authors: | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
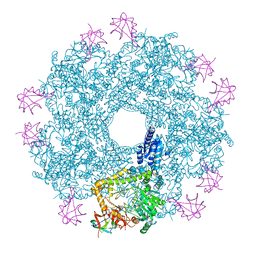
5FKZ
 
 | | Structure of E.coli Constitutive lysine decarboxylase | | Descriptor: | LYSINE DECARBOXYLASE, CONSTITUTIVE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
7K08
 
 | |
1LVN
 
 | |
5OHG
 
 | | enolase in complex with RNase E | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Du, D, Luisi, B.F. | | Deposit date: | 2017-07-16 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes.
Nucleic Acids Res., 46, 2018
|
|
4N7U
 
 | |
