6N58
 
 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound with TraR in conformation II | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-21 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
6NQ6
 
 | | Structure & function of a new Aspartylglucosaminuria variant | | Descriptor: | N(4)-(Beta-N-acetylglucosaminyl)-L-asparaginase | | Authors: | Pande, S, Guo, H.C. | | Deposit date: | 2019-01-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The T99K variant of glycosylasparaginase shows a new structural mechanism of the genetic disease aspartylglucosaminuria.
Protein Sci., 28, 2019
|
|
6OUL
 
 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound to rpsTP2 promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-05-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
6QXA
 
 | | Structure of membrane bound pyrophosphatase from Thermotoga maritima in complex with imidodiphosphate and N-[(2-amino-6-benzothiazolyl)methyl]-1H-indole-2-carboxamide (ATC) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, IMIDODIPHOSPHORIC ACID, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Vidilaseris, K, Goldman, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Asymmetry in catalysis byThermotoga maritimamembrane-bound pyrophosphatase demonstrated by a nonphosphorus allosteric inhibitor.
Sci Adv, 5, 2019
|
|
7EP8
 
 | |
7C2N
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SPIRO | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1'-(2,3-dihydro-1,4-benzodioxin-6-ylmethyl)spiro[6,7-dihydrothieno[3,2-c]pyran-4,4'-piperidine], Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7C2M
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with NITD-349 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Chimera of drug exporters of the RND superfamily-like protein and Endolysin, N-(4,4-dimethylcyclohexyl)-4,6-bis(fluoranyl)-1H-indole-2-carboxamide, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
6V5B
 
 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - Active state | | Descriptor: | CALCIUM ION, Microprocessor complex subunit DGCR8, Pri-miR-16-2 (78-MER), ... | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
8B8T
 
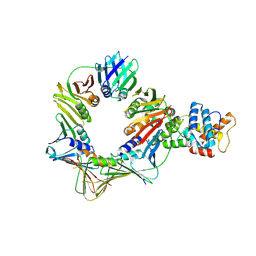 | | Open conformation of the complex of DNA ligase I on PCNA and DNA in the presence of ATP | | Descriptor: | DNA ligase 1, Proliferating cell nuclear antigen | | Authors: | Blair, K, Tehseen, M, Raducanu, V.S, Shahid, T, Lancey, C, Cruehet, R, Hamdan, S, De Biasio, A. | | Deposit date: | 2022-10-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of human Lig1 regulation by PCNA in Okazaki fragment sealing.
Nat Commun, 13, 2022
|
|
6VLB
 
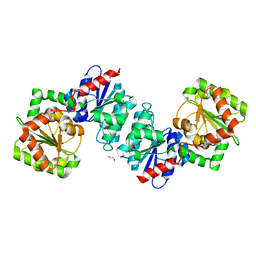 | |
6VLC
 
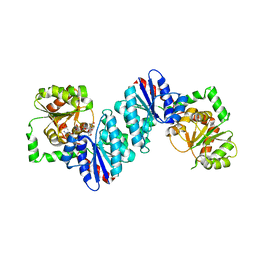 | |
6W54
 
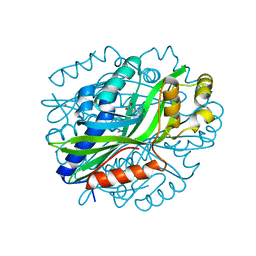 | | Crystal Structure of Gallic Acid Decarboxylase from Arxula adeninivorans | | Descriptor: | 4-NITROCATECHOL, COBALT (II) ION, Gallate decarboxylase, ... | | Authors: | Zeug, M, Marckovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
8CH6
 
 | | Structure of a late-stage activated spliceosome (BAqr) arrested with a dominant-negative Aquarius mutant (state B complex). | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Cretu, C, Schmitzova, J, Pena, V. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of catalytic activation in human splicing.
Nature, 617, 2023
|
|
8C0D
 
 | | UFL1/DDRGK1 bound to UFC1 | | Descriptor: | DDRGK domain-containing protein 1, E3 UFM1-protein ligase 1, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Magnussen, H.M, Kulathu, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.564 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
8BZR
 
 | | UFC1-UFM1 conjugate | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin-fold modifier 1, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Magnussen, H.M, Kulathu, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
8D49
 
 | | Structure of Cas12a2 binary complex | | Descriptor: | OrfB_Zn_ribbon domain-containing protein, RNA (26-MER) | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2022-06-01 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | RNA targeting unleashes indiscriminate nuclease activity of CRISPR-Cas12a2.
Nature, 613, 2023
|
|
8D4A
 
 | | Cas12a2 quaternary complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2022-06-01 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | RNA targeting unleashes indiscriminate nuclease activity of CRISPR-Cas12a2.
Nature, 613, 2023
|
|
8D4B
 
 | | Structure of Cas12a2 ternary complex | | Descriptor: | OrfB_Zn_ribbon domain-containing protein, RNA (28-MER), RNA (41-MER) | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2022-06-01 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | RNA targeting unleashes indiscriminate nuclease activity of CRISPR-Cas12a2.
Nature, 613, 2023
|
|
1JZC
 
 | |
8H3U
 
 | | Inhibitor-bound EP, polyA model | | Descriptor: | Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-10-09 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
8H3S
 
 | | Substrate-bound EP, polyA model | | Descriptor: | Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain, Serine protease 1 | | Authors: | Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-10-09 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
8HG1
 
 | | The structure of MPXV polymerase holoenzyme in replicating state | | Descriptor: | DNA (25-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Peng, Q, Xie, Y.F, Kuai, L, Wang, H, Qi, J.X, Gao, F, Shi, Y. | | Deposit date: | 2022-11-13 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of monkeypox virus DNA polymerase holoenzyme.
Science, 379, 2023
|
|
5UJM
 
 | | Structure of the active form of human Origin Recognition Complex and its ATPase motor module | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Tocilj, A, On, K, Yuan, Z, Sun, J, Elkayam, E, Li, H, Stillman, B, Joshua-Tor, L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structure of the active form of human Origin Recognition Complex and its ATPase motor module.
Elife, 6, 2017
|
|
5T63
 
 | | The HhoA protease from Synechocystis sp. PCC 6803 | | Descriptor: | ALA-ALA-ALA-ALA, MAGNESIUM ION, Putative serine protease HhoA | | Authors: | Persson, K, Hall, M, Funk, C. | | Deposit date: | 2016-09-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The HhoA protease from Synechocystis sp. PCC 6803 - Novel insights into structure and activity regulation.
J. Struct. Biol., 198, 2017
|
|
5T69
 
 | | The HhoA protease from Synechocystis sp. PCC 6803, active site mutant | | Descriptor: | ACETATE ION, MAGNESIUM ION, Putative serine protease HhoA | | Authors: | Persson, K, Hall, M, Funk, C. | | Deposit date: | 2016-09-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The HhoA protease from Synechocystis sp. PCC 6803 - Novel insights into structure and activity regulation.
J. Struct. Biol., 198, 2017
|
|
