4CTC
 
 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 7-amino-3-cyclopropyl-12-fluoro-1,10,16-trimethyl-16,17- dihydro-1H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecin-15(10H)-one | | Descriptor: | (10R)-7-amino-3-cyclopropyl-12-fluoro-1,10,16-trimethyl-16,17-dihydro-1H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecin-15(10H)-one, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
1BSD
 
 | |
1BNG
 
 | | BARNASE S85C/H102C DISULFIDE MUTANT | | Descriptor: | BARNASE | | Authors: | Clarke, J, Henrick, K, Fersht, A.R. | | Deposit date: | 1995-03-31 | | Release date: | 1995-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Disulfide mutants of barnase. I: Changes in stability and structure assessed by biophysical methods and X-ray crystallography.
J.Mol.Biol., 253, 1995
|
|
1BSB
 
 | |
8R1O
 
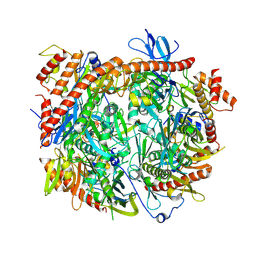 | | Structure of C. thermophilum RNA exosome core | | Descriptor: | Exoribonuclease phosphorolytic domain-containing protein, Exoribonuclease-like protein, Exosome complex component MTR3, ... | | Authors: | Lazzaretti, D, Liebau, J, Pilsl, M, Sprangers, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Beyond static structures: quantitative dynamics in the eukaryotic RNA exosome complex
Biorxiv, 2024
|
|
8XOM
 
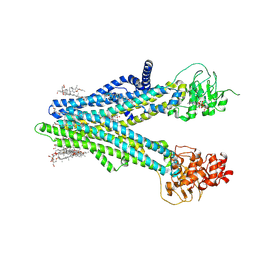 | | Cryo-EM structure of human ABCC4 in complex with ANP-bound in NBD1 and METHOTREXATE | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, MAGNESIUM ION, ... | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 2024
|
|
8XOL
 
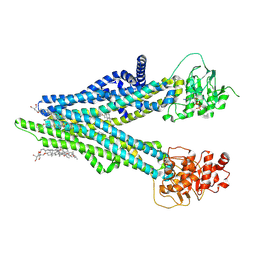 | | Cryo-EM structure of human ABCC4 with ANP bound in NBD1 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, MAGNESIUM ION, ... | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 2024
|
|
1S02
 
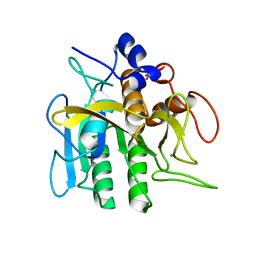 | | EFFECTS OF ENGINEERED SALT BRIDGES ON THE STABILITY OF SUBTILISIN BPN' | | Descriptor: | CALCIUM ION, SUBTILISIN BPN', SULFATE ION | | Authors: | Erwin, C.R, Barnett, B.L, Oliver, J.D, Sullivan, J.F. | | Deposit date: | 1991-02-20 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effects of engineered salt bridges on the stability of subtilisin BPN'.
Protein Eng., 4, 1990
|
|
8TAV
 
 | |
4DD8
 
 | | ADAM-8 metalloproteinase domain with bound batimastat | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hall, T, Shieh, H.S, Day, J.E, Caspers, N, Chrencik, J.E, Williams, J.M, Pegg, L.E, Pauley, A.M, Moon, A.F, Krahn, J.M, Fischer, D.H, Kiefer, J.R, Tomasselli, A.G, Zack, M.D. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human ADAM-8 catalytic domain complexed with batimastat.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
169L
 
 | |
178L
 
 | | Protein flexibility and adaptability seen in 25 crystal forms of T4 LYSOZYME | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Matsumura, M, Weaver, L, Zhang, X.-J, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
4E0M
 
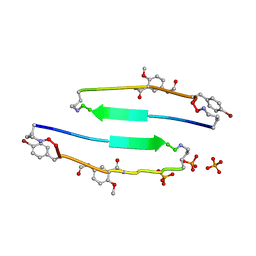 | | SVQIVYK segment from human Tau (305-311) displayed on 54-membered macrocycle scaffold (form I) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide SVQIVYK(ORN)EF(HAO)(4BF)K(ORN), PHOSPHATE ION | | Authors: | Zhao, M, Liu, C, Michael, S.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway
to toxic amyloid aggregates
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2VOY
 
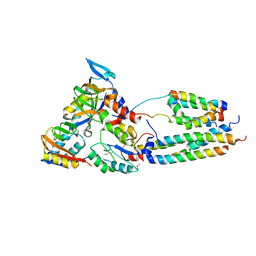 | | CryoEM model of CopA, the copper transporting ATPase from Archaeoglobus fulgidus | | Descriptor: | CATION-TRANSPORTING ATPASE, P-TYPE, POTENTIAL COPPER-TRANSPORTING ATPASE, ... | | Authors: | Wu, C.-C, Rice, W.J, Stokes, D.L. | | Deposit date: | 2008-02-25 | | Release date: | 2009-05-26 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structure of a Copper Pump Suggests a Regulatory Role for its Metal-Binding Domain.
Structure, 16, 2008
|
|
1U3U
 
 | |
4E0L
 
 | | FYLLYYT segment from human Beta 2 Microglobulin (62-68) displayed on 54-membered macrocycle scaffold | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide FYLLYYT(ORN)KN(HAO)SA(ORN) | | Authors: | Zhao, M, Liu, C, Michael, S.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WQ2
 
 | |
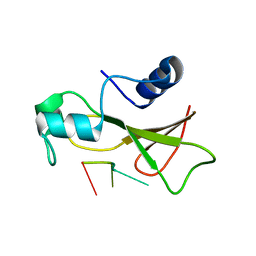
1BRN
 
 | |
1BSC
 
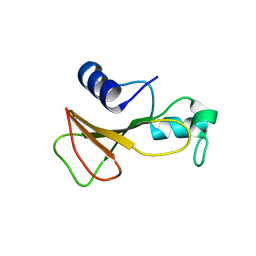 | |
1C4D
 
 | | GRAMICIDIN CSCL COMPLEX | | Descriptor: | CESIUM ION, CHLORIDE ION, GRAMICIDIN A, ... | | Authors: | Wallace, B.A. | | Deposit date: | 1999-06-04 | | Release date: | 2000-01-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Gramicidin Pore: Crystal Structure of a Cesium Complex.
Science, 241, 1988
|
|
1U3W
 
 | |
2X4N
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to residual fragments of a photocleavable peptide that is cleaved upon UV-light treatment | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
1AQN
 
 | | SUBTILISIN MUTANT 8324 | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SUBTILISIN 8324, ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-07-31 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stabilityfor subtilisin BPN' through incremental changes in the free energy of unfolding
To be published
|
|
4JD2
 
 | |
1BAO
 
 | |
