1YIY
 
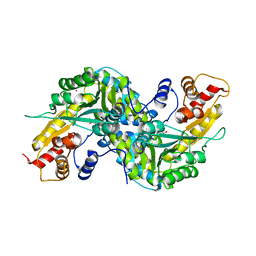 | | Aedes aegypti kynurenine aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BROMIDE ION, kynurenine aminotransferase; glutamine transaminase K | | Authors: | Han, Q, Gao, Y.G, Robinson, H, Ding, H, Wilson, S, Li, J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Aedes aegypti kynurenine aminotransferase.
FEBS J., 272, 2005
|
|
1YIR
 
 | |
1KAX
 
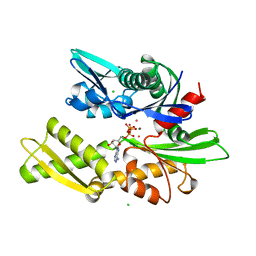 | | 70KD HEAT SHOCK COGNATE PROTEIN ATPASE DOMAIN, K71M MUTANT | | Descriptor: | 70KD HEAT SHOCK COGNATE PROTEIN, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | O'Brien, M.C, Flaherty, K.M, Mckay, D.B. | | Deposit date: | 1996-04-15 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lysine 71 of the chaperone protein Hsc70 Is essential for ATP hydrolysis.
J.Biol.Chem., 271, 1996
|
|
1Y8Q
 
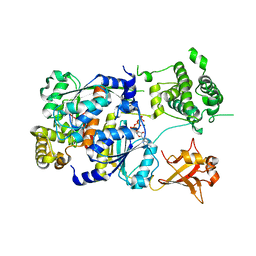 | | SUMO E1 ACTIVATING ENZYME SAE1-SAE2-MG-ATP COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ubiquitin-like 1 activating enzyme E1A, ... | | Authors: | Lois, L.M, Lima, C.D. | | Deposit date: | 2004-12-13 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1
Embo J., 24, 2005
|
|
1YIZ
 
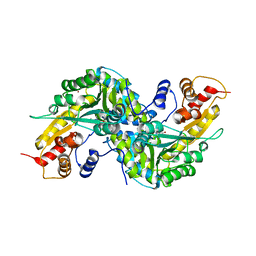 | | Aedes aegypti kynurenine aminotrasferase | | Descriptor: | BROMIDE ION, kynurenine aminotransferase; glutamine transaminase | | Authors: | Han, Q, Gao, Y.G, Robinson, H, Ding, H, Wilson, S, Li, J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of Aedes aegypti kynurenine aminotransferase.
FEBS J., 272, 2005
|
|
1KNY
 
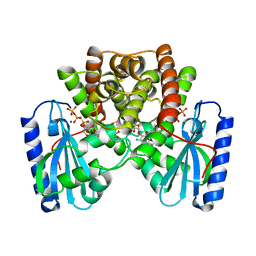 | | KANAMYCIN NUCLEOTIDYLTRANSFERASE | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, KANAMYCIN A, KANAMYCIN NUCLEOTIDYLTRANSFERASE, ... | | Authors: | Pedersen, L.C, Benning, M.M, Holden, H.M. | | Deposit date: | 1995-07-07 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase.
Biochemistry, 34, 1995
|
|
1YJ0
 
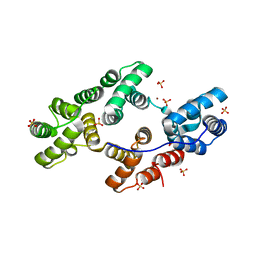 | | Crystal Structures of Chicken Annexin V in Complex with Zn2+ | | Descriptor: | Annexin A5, SULFATE ION, ZINC ION | | Authors: | Ortlund, E.A, Chai, G, Genge, B, Wu, L.N.Y, Wuthier, R.E, Lebioda, L. | | Deposit date: | 2005-01-11 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structures of Chicken Annexin A5 in Complex with Functional Modifiers Ca2+ and Zn2+ Reveal Zn2+ Induced Formation of Non-Planar Assemblies
Annexins, 1, 2005
|
|
1KAZ
 
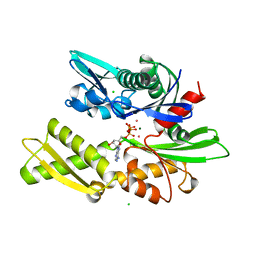 | | 70KD HEAT SHOCK COGNATE PROTEIN ATPASE DOMAIN, K71E MUTANT | | Descriptor: | 70KD HEAT SHOCK COGNATE PROTEIN, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | O'Brien, M.C, Flaherty, K.M, Mckay, D.B. | | Deposit date: | 1996-04-15 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lysine 71 of the chaperone protein Hsc70 Is essential for ATP hydrolysis.
J.Biol.Chem., 271, 1996
|
|
1YJ1
 
 | | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ubiquitin | | Authors: | Bang, D, Makhatadze, G.I, Tereshko, V, Kossiakoff, A.A, Kent, S.B. | | Deposit date: | 2005-01-13 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YJ2
 
 | | Cyclized, non-dehydrated post-translational product for S65A Y66S H148G GFP variant | | Descriptor: | 1,2-ETHANEDIOL, Green Fluorescent Protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-01-13 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Understanding GFP Chromophore Biosynthesis: Controlling Backbone Cyclization and Modifying Post-translational Chemistry.
Biochemistry, 44, 2005
|
|
4CJ8
 
 | | monoclinic crystal form of Bogt6a E192Q in complex with UDP-GalNAc, UDP and GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, GLYCEROL, ... | | Authors: | Pham, T, Stinson, B, Thiyagarajan, N, Lizotte-Waniewski, M, Brew, K, Acharya, K.R. | | Deposit date: | 2013-12-19 | | Release date: | 2014-02-05 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of Complexes of a Metal-Independent Glycosyltransferase Gt6 from Bacteroides Ovatus with Udp-Galnac and its Hydrolysis Products
J.Biol.Chem., 289, 2014
|
|
1Y9W
 
 | | Structural Genomics, 1.9A crystal structure of an acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyltransferase | | Authors: | Zhang, R, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-16 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9A crystal structure of an acetyltransferase from Bacillus cereus ATCC 14579
To be Published, 2005
|
|
1Y9Z
 
 | |
1YAZ
 
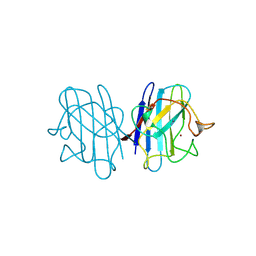 | | AZIDE-BOUND YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | AZIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 2000-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1YJ3
 
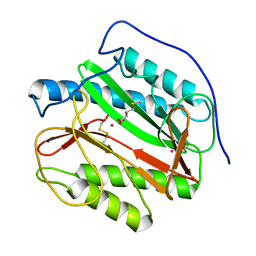 | | Crystal structure analysis of product bound methionine aminopeptidase Type 1c from Mycobacterium Tuberculosis | | Descriptor: | BETA-MERCAPTOETHANOL, COBALT (II) ION, METHIONINE, ... | | Authors: | Addlagatta, A, Quillin, M.L, Omotoso, O, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-01-13 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of an SH3-binding motif in a new class of methionine aminopeptidases from Mycobacterium tuberculosis suggests a mode of interaction with the ribosome.
Biochemistry, 44, 2005
|
|
1YB7
 
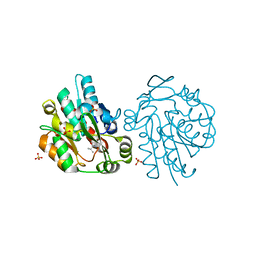 | | Hydroxynitrile lyase from hevea brasiliensis in complex with 2,3-dimethyl-2-hydroxy-butyronitrile | | Descriptor: | (S)-2-HYDROXY-2,3-DIMETHYLBUTANENITRILE, (S)-acetone-cyanohydrin lyase, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Kratky, C. | | Deposit date: | 2004-12-20 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural determinants of the enantioselectivity of the hydroxynitrile lyase from Hevea brasiliensis
J.Biotechnol., 129, 2007
|
|
1YBG
 
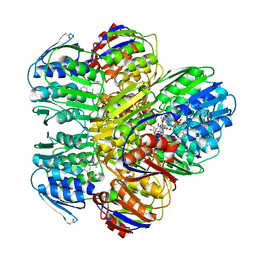 | | MurA inhibited by a derivative of 5-sulfonoxy-anthranilic acid | | Descriptor: | N-METHYL-N-{2-[(2-NAPHTHYLSULFONYL)AMINO]-5-[(2-NAPHTHYLSULFONYL)OXY]BENZOYL}-L-ASPARTIC ACID, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Eschenburg, S, Priestman, M.A, Abdul-Latif, F.A, Delachaume, C, Fassy, F, Schonbrunn, E. | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Inhibitor That Suspends the Induced Fit Mechanism of UDP-N-acetylglucosamine Enolpyruvyl Transferase (MurA).
J.Biol.Chem., 280, 2005
|
|
1YJ4
 
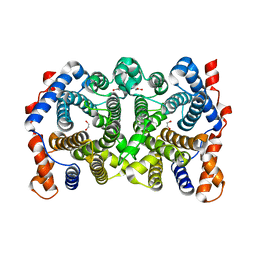 | | Y305F Trichodiene Synthase | | Descriptor: | 1,2-ETHANEDIOL, Trichodiene synthase | | Authors: | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-01-13 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
1KJS
 
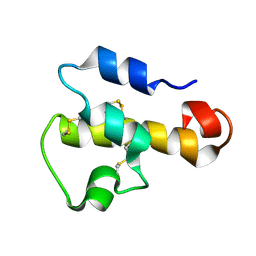 | | NMR SOLUTION STRUCTURE OF C5A AT PH 5.2, 303K, 20 STRUCTURES | | Descriptor: | C5A | | Authors: | Zhang, X, Boyar, W, Toth, M, Wennogle, L, Gonnella, N.C. | | Deposit date: | 1997-01-09 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structural definition of the C5a C terminus by two-dimensional nuclear magnetic resonance spectroscopy.
Proteins, 28, 1997
|
|
4CTH
 
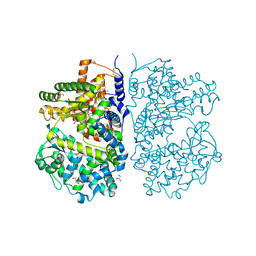 | | Neprilysin variant G399V,G714K in complex with phosphoramidon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Webster, C.I, Burrell, M, Olsson, L, Fowler, S.B, Digby, S, Sandercock, A, Snijder, A, Tebbe, J, Haupts, U, Grudzinska, J, Jermutus, L, Andersson, C. | | Deposit date: | 2014-03-13 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering Neprilysin Activity and Specificity to Create a Novel Therapeutic for Alzheimer'S Disease.
Plos One, 9, 2014
|
|
1YJ6
 
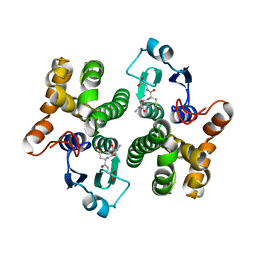 | |
1YHO
 
 | | Solution structure of human dihydrofolate reductase complexed with trimethoprim and nadph, 25 structures | | Descriptor: | 2,4-DIAMINO-5-(3,4,5-TRIMETHOXY-BENZYL)-PYRIMIDIN-1-IUM, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Polshakov, V.I, Birdsall, B. | | Deposit date: | 2005-01-10 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human dihydrofolate reductase in its complex with trimethoprim and NADPH.
J.Biomol.Nmr, 33, 2005
|
|
1YJ7
 
 | | Crystal structure of enteropathogenic E.coli (EPEC) type III secretion system protein EscJ | | Descriptor: | GLYCEROL, PHOSPHATE ION, escJ | | Authors: | Yip, C.K, Kimbrough, T.G, Felise, H.B, Vuckovic, M, Thomas, N.A, Pfuetzner, R.A, Frey, E.A, Finlay, B.B, Miller, S.I, Strynadka, N.C.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the molecular platform for type III secretion system assembly.
Nature, 435, 2005
|
|
1YC2
 
 | | Sir2Af2-NAD-ADPribose-nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase 2, ... | | Authors: | Avalos, J.L, Bever, M.K, Wolberger, C. | | Deposit date: | 2004-12-21 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Sirtuin Inhibition by Nicotinamide: Altering the NAD(+) Cosubstrate Specificity of a Sir2 Enzyme.
Mol.Cell, 17, 2005
|
|
1YD6
 
 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Bacillus caldotenax | | Descriptor: | CHLORIDE ION, SULFATE ION, UvrC | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
