4XBB
 
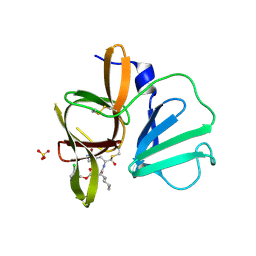 | | 1.85A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor diethyl [(1R,2S)-2-[(N-{[(3-chlorobenzyl)oxy]carbonyl}-3-cyclohexyl-L-alanyl)amino]-1-hydroxy-3-(2-oxo-2H-pyrrol-3-yl)propyl]phosphonate | | Descriptor: | 3C-LIKE PROTEASE, SULFATE ION, diethyl [(1R,2S)-2-[(N-{[(3-chlorobenzyl)oxy]carbonyl}-3-cyclohexyl-L-alanyl)amino]-1-hydroxy-3-(2-oxo-2H-pyrrol-3-yl)propyl]phosphonate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
5XVK
 
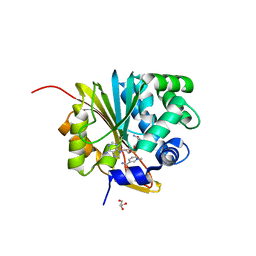 | | Crystal structure of mouse Nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl Nicotinamide (MNA) | | Descriptor: | 3-carbamoyl-1-methylpyridin-1-ium, GLYCEROL, Nicotinamide N-methyltransferase, ... | | Authors: | Swaminathan, S, Birudukota, S, Thakur, M.K, Parveen, R, Kandan, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of monkey and mouse nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl nicotinamide
Biochem. Biophys. Res. Commun., 491, 2017
|
|
5XRW
 
 | |
5XVR
 
 | |
5XS3
 
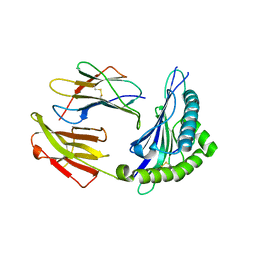 | | Crystal structure of HLA Class I antigen | | Descriptor: | Heavy Chain, Light Chain, P | | Authors: | Wei, P.C, Yang, Y, Liu, Z.X, Luo, Z.Q, Tu, W.Y, Han, J.Y, Deng, Y.H, Yin, L. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of Autoantigen Presentation by HLA-C*06:02 in Psoriasis
J. Invest. Dermatol., 137, 2017
|
|
4XOR
 
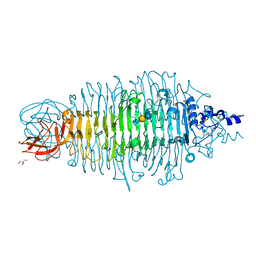 | | Tailspike protein double mutant D339N/E372Q of E. coli bacteriophage HK620 in complex with pentasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
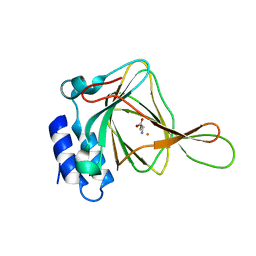
4XFH
 
 | |
4XHC
 
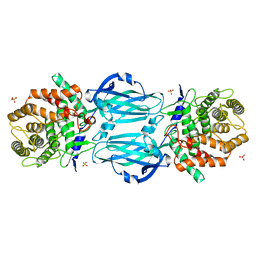 | | rhamnosidase from Klebsiella oxytoca with rhamnose bound | | Descriptor: | Alpha-L-rhamnosidase, SULFATE ION, alpha-L-rhamnopyranose | | Authors: | O'Neill, E.O, Stevenson, C.E.M, Patterson, M.J, Rejzek, M, Chauvin, A, Lawson, D.M, Field, R.A. | | Deposit date: | 2015-01-05 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a novel two domain GH78 family alpha-rhamnosidase from Klebsiella oxytoca with rhamnose bound.
Proteins, 83, 2015
|
|
5XUU
 
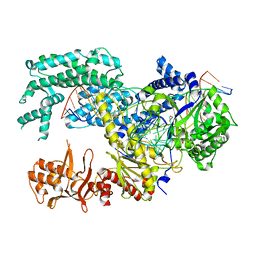 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*CP*CP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
5XW2
 
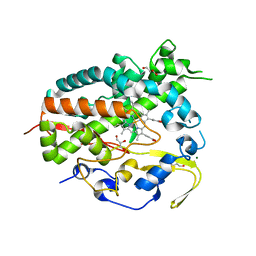 | |
5XVP
 
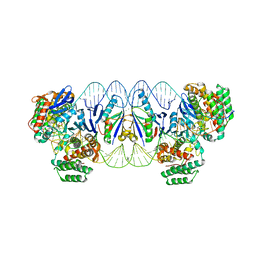 | | E. fae Cas1-Cas2/prespacer/target ternary complex revealing the fully integrated states | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(P*TP*TP*CP*TP*CP*CP*GP*AP*G)-3'), ... | | Authors: | Xiao, Y, Ng, S, Nam, K.H, Ke, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration.
Nature, 550, 2017
|
|
4XLC
 
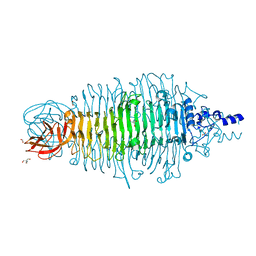 | | Tailspike protein double mutant D339N/E372A of E. coli bacteriophage HK620 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-13 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
5X87
 
 | |
4XOQ
 
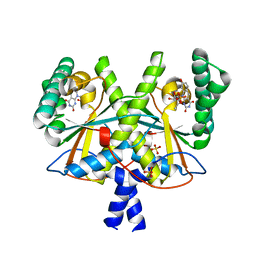 | | F420 complex of coenzyme F420:L-glutamate ligase (FbiB) from Mycobacterium tuberculosis (C-terminal domain) | | Descriptor: | COENZYME F420, Coenzyme F420:L-glutamate ligase, SULFATE ION | | Authors: | Rehan, A.M, Bashiri, G, Baker, H.M, Baker, E.N, Squire, C.J. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Elongation of the Poly-gamma-glutamate Tail of F420 Requires Both Domains of the F420: gamma-Glutamyl Ligase (FbiB) of Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
4XDE
 
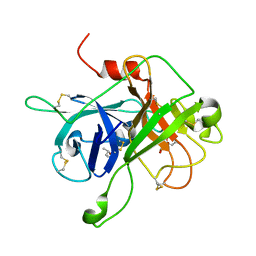 | | Coagulation Factor XII protease domain crystal structure | | Descriptor: | CITRATE ANION, Coagulation factor XII, ISOPROPYL ALCOHOL | | Authors: | Pathak, M, Wilmann, P, Awford, J, Li, C, Fisher, P.M, Dreveny, I, Dekker, L.V, Emsley, J. | | Deposit date: | 2014-12-19 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Coagulation factor XII protease domain crystal structure.
J.Thromb.Haemost., 13, 2015
|
|
4XDO
 
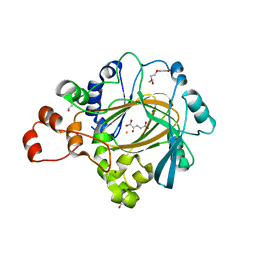 | | Crystal structure of human KDM4C catalytic domain with OGA | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Lysine-specific demethylase 4C, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A High-Throughput Mass Spectrometry Assay Coupled with Redox Activity Testing Reduces Artifacts and False Positives in Lysine Demethylase Screening.
J Biomol Screen, 20, 2015
|
|
4XCI
 
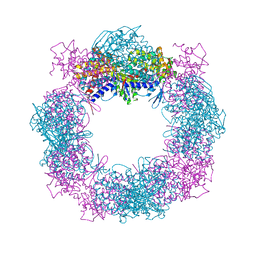 | | Crystal structure of a hexadecameric TF55 complex from S. solfataricus, crystal form II | | Descriptor: | Thermosome subunit alpha, Thermosome subunit beta | | Authors: | Stewart, A.G, Chaston, J.J, Smits, C, Stock, D. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0023 Å) | | Cite: | Structural and Functional Insights into the Evolution and Stress Adaptation of Type II Chaperonins.
Structure, 24, 2016
|
|
5Y3Z
 
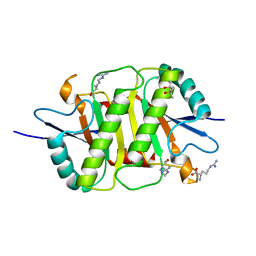 | | Structure of the periplasmic domain of the MotB L119P mutant from Salmonella (crystal form 1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ARGININE, IMIDAZOLE, ... | | Authors: | Takao, M, Kojima, S, Sakuma, M, Homma, M, Imada, K. | | Deposit date: | 2017-07-31 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Helix Rearrangement in the Periplasmic Domain of the Flagellar Stator B Subunit Activates Peptidoglycan Binding and Ion Influx.
Structure, 26, 2018
|
|
4XD5
 
 | | Phosphotriesterase variant R2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R2, ... | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
5Y46
 
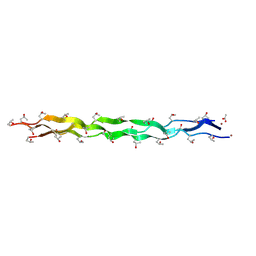 | |
4XFB
 
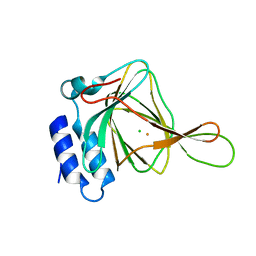 | | cysteine dioxygenase variant - C93A at pH 8.0 unliganded | | Descriptor: | CHLORIDE ION, Cysteine dioxygenase type 1, FE (III) ION | | Authors: | Driggers, C.M, Karplus, P.A. | | Deposit date: | 2014-12-26 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Insights into the Role of the Cys-Tyr Crosslink and Inhibitor Recognition by Mammalian Cysteine Dioxygenase.
J. Mol. Biol., 428, 2016
|
|
4XG0
 
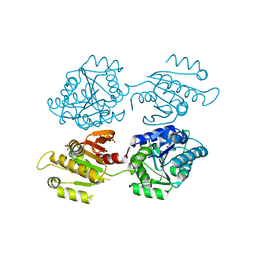 | | Crystal structure of a domain of unknown function (DUF1537) from Bordetella bronchiseptica (BB3215), Target EFI-511620, with bound citrate, domain swapped dimer, space group C2221 | | Descriptor: | CHLORIDE ION, CITRIC ACID, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-30 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y3J
 
 | | Crystal structure of horse TLR9 in complex with two DNAs (CpG DNA and TCGCAC DNA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*AP*GP*GP*CP*GP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2017-07-29 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Toll-like Receptor 9 Contains Two DNA Binding Sites that Function Cooperatively to Promote Receptor Dimerization and Activation
Immunity, 48, 2018
|
|
4XLF
 
 | | Tailspike protein double mutant D339N/E372A of E. coli bacteriophage HK620 in complex with pentasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-13 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
