1YMH
 
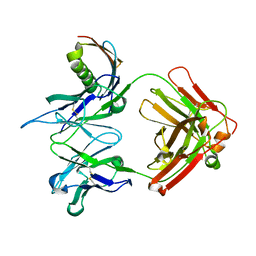 | | anti-HCV Fab 19D9D6 complexed with protein L (PpL) mutant A66W | | Descriptor: | Fab 16D9D6, heavy chain, light chain, ... | | Authors: | Granata, V, Housden, N.G, Harrison, S, Jolivet-Reynaud, C, Gore, M.G, Stura, E.A. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparison of the crystallization and crystal packing of two Fab single-site mutant protein L complexes.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2KC0
 
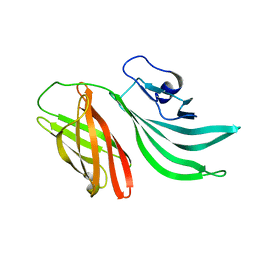 | | Solution structure of the factor H binding protein | | Descriptor: | lipoprotein | | Authors: | Cantini, F, Veggi, D, Dragonetti, S, Savino, S, Scarselli, M, Romagnoli, G, Pizza, M, Banci, L, Rappuoli, R. | | Deposit date: | 2008-12-13 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Factor H-binding Protein, a Survival Factor and Protective Antigen of Neisseria meningitidis
J.Biol.Chem., 284, 2009
|
|
4CD1
 
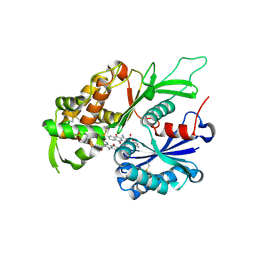 | | RnNTPDase2 in complex with PSB-071 | | Descriptor: | 1-AMINO-4-(3-METHYLPHENYL)AMINO-9,10-DIOXO-9,10-DIHYDROANTHRACENE-2-SULFONATE, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2 | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2013-10-29 | | Release date: | 2014-02-12 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Ntpdase2 in Complex with the Sulfoanthraquinone Inhibitor Psb-071.
J.Struct.Biol., 185, 2014
|
|
4CD3
 
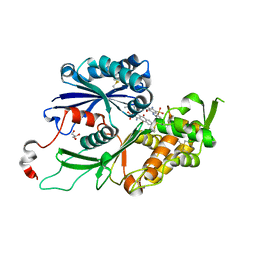 | | RnNTPDase2 X4 variant in complex with PSB-071 | | Descriptor: | 1-AMINO-4-(3-METHYLPHENYL)AMINO-9,10-DIOXO-9,10-DIHYDROANTHRACENE-2-SULFONATE, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, GLYCEROL | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2013-10-29 | | Release date: | 2014-02-12 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Ntpdase2 in Complex with the Sulfoanthraquinone Inhibitor Psb-071.
J.Struct.Biol., 185, 2014
|
|
1XKG
 
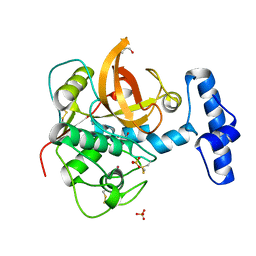 | | Crystal structure of the major house dust mite allergen Der p 1 in its pro form at 1.61 A resolution | | Descriptor: | GLYCEROL, Major mite fecal allergen Der p 1, SULFATE ION, ... | | Authors: | Meno, K, Thorsted, P.B, Gajhede, M. | | Deposit date: | 2004-09-29 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The crystal structure of recombinant proDer p 1, a major house dust mite proteolytic allergen.
J.Immunol., 175, 2005
|
|
1Y12
 
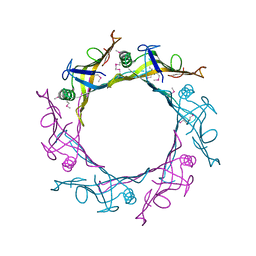 | |
4EC6
 
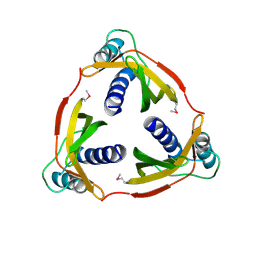 | | Ntf2-like, potential transfer protein TraM from Gram-positive conjugative plasmid pIP501 | | Descriptor: | Putative uncharacterized protein | | Authors: | Goessweiner-Mohr, N, Grumet, L, Pavkov-Keller, T, Wang, M, Keller, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-12-05 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A Structure of the Enterococcus Conjugation Protein TraM resembles VirB8 Type IV Secretion Proteins.
J.Biol.Chem., 288, 2013
|
|
4DRR
 
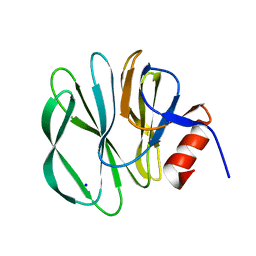 | | Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen | | Descriptor: | Outer capsid protein VP4, SODIUM ION | | Authors: | Hu, L, Crawford, S.E, Czako, R, Cortes-Penfield, N.W, Smith, D.F, Le Pendu, J, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen.
Nature, 485, 2012
|
|
4DS0
 
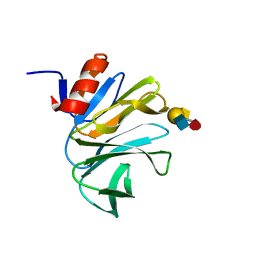 | | Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen | | Descriptor: | Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hu, L, Crawford, S.E, Czako, R, Cortes-Penfield, N.W, Smith, D.F, Le Pendu, J, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen.
Nature, 485, 2012
|
|
4DRV
 
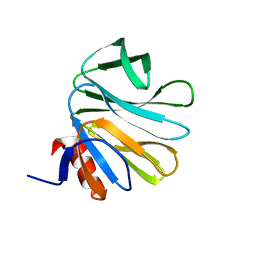 | | Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen | | Descriptor: | Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Hu, L, Crawford, S.E, Czako, R, Cortes-Penfield, N.W, Smith, D.F, Le Pendu, J, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen.
Nature, 485, 2012
|
|
2CYG
 
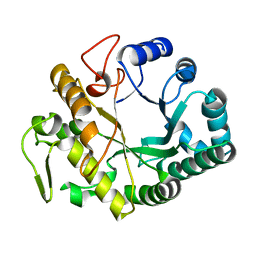 | | Crystal structure at 1.45- resolution of the major allergen endo-beta-1,3-glucanase of banana as a molecular basis for the latex-fruit syndrome | | Descriptor: | beta-1, 3-glucananse | | Authors: | Receveur-Brechot, V, Czjzek, M, Barre, A, Roussel, A, Peumans, W.J, Van Damme, E.J.M, Rouge, P. | | Deposit date: | 2005-07-07 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure at 1.45-A resolution of the major allergen endo-beta-1,3-glucanase of banana as a molecular basis for the latex-fruit syndrome
Proteins, 63, 2006
|
|
4EEF
 
 | |
2DF7
 
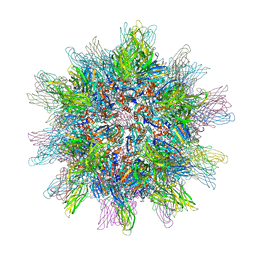 | | Crystal structure of infectious bursal disease virus VP2 subviral particle | | Descriptor: | CALCIUM ION, CHLORIDE ION, structural polyprotein VP2 | | Authors: | Ko, T.P, Lee, C.C, Wang, M.Y, Wang, A.H. | | Deposit date: | 2006-02-27 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of infectious bursal disease virus VP2 subviral particle at 2.6A resolution: Implications in virion assembly and immunogenicity.
J.Struct.Biol., 155, 2006
|
|
2JDG
 
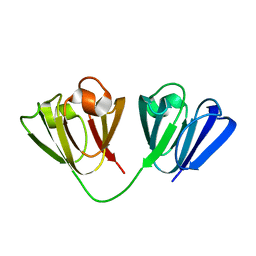 | | Affilin based on HUMAN GAMMA-B CRYSTALLIN | | Descriptor: | GAMMA CRYSTALLIN B | | Authors: | Ebersbach, H, Fiedler, E, Scheuermann, T, Fiedler, M, Stubbs, M.T, Reimann, C, Proetzel, G, Rudolph, R, Fiedler, U. | | Deposit date: | 2007-01-08 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affilin-Novel Binding Molecules Based on Human Gamma-B-Crystallin, an All Beta-Sheet Protein.
J.Mol.Biol., 372, 2007
|
|
2LS0
 
 | |
2KZI
 
 | | Solution structure of the ZHER2 Affibody | | Descriptor: | Engineered protein, ZHER2 Affibody | | Authors: | Hard, T. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity HER2 receptor binding by an engineered protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2FLG
 
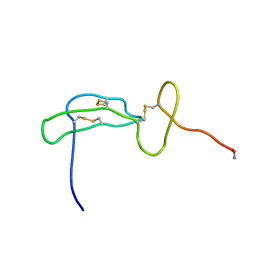 | | Solution structure of an EGF-LIKE domain from the Plasmodium falciparum merozoite surface protein 1 | | Descriptor: | Merozoite surface protein 1 | | Authors: | James, S, Moehle, K, Pluschke, G, Robinson, J. | | Deposit date: | 2006-01-06 | | Release date: | 2006-02-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Synthesis, solution structure and immune recognition of an epidermal growth factor-like domain from Plasmodium falciparum merozoite surface protein-1.
Chembiochem, 7, 2006
|
|
2KZJ
 
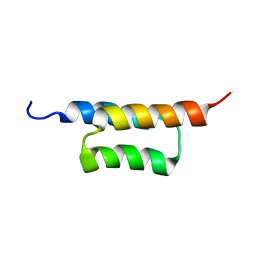 | | Solution structure of the ZHER2 Affibody (alternative) | | Descriptor: | Engineered protein, ZHER2 Affibody | | Authors: | Hard, T. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity HER2 receptor binding by an engineered protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2LNL
 
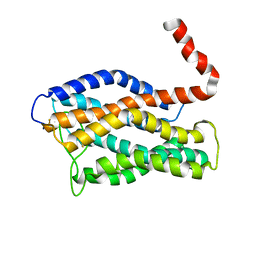 | | Structure of human CXCR1 in phospholipid bilayers | | Descriptor: | C-X-C chemokine receptor type 1 | | Authors: | Park, S, Das, B.B, Casagrande, F, Nothnagel, H, Chu, M, Kiefer, H, Maier, K, De Angelis, A, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-12-31 | | Release date: | 2012-10-17 | | Last modified: | 2016-04-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the chemokine receptor CXCR1 in phospholipid bilayers.
Nature, 491, 2012
|
|
5LVC
 
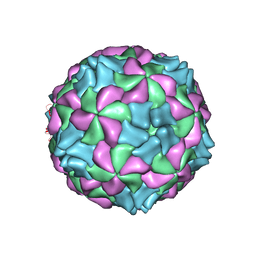 | | Aichi virus 1: empty particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Sabin, C, Fuzik, T, Skubnik, K, Palkova, L, Lindberg, A.M, Plevka, P. | | Deposit date: | 2016-09-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Aichi Virus 1 and Its Empty Particle: Clues to Kobuvirus Genome Release Mechanism.
J.Virol., 90, 2016
|
|
2LM3
 
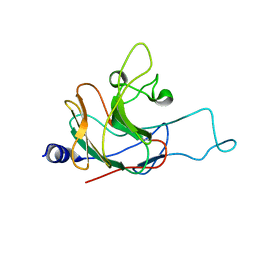 | | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain | | Descriptor: | Tripartite motif-containing protein 5 | | Authors: | Biris, N, Yang, Y, Taylor, A.B, Tomashevski, A, Guo, M, Hart, P.J, Diaz-Griffero, F, Ivanov, D.N. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1OUV
 
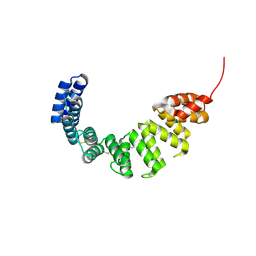 | | Helicobacter cysteine rich protein C (HcpC) | | Descriptor: | conserved hypothetical secreted protein | | Authors: | Mittl, P.R, Luethy, L. | | Deposit date: | 2003-03-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Helicobacter Cysteine-rich Protein C at 2.0A Resolution: Similar Peptide-binding Sites in TPR and SEL1-like Repeat Proteins
J.Mol.Biol., 340, 2004
|
|
6RJ1
 
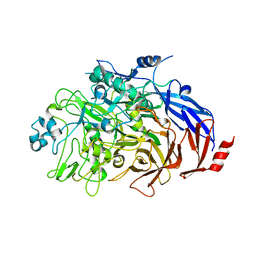 | |
2ME4
 
 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2ME1
 
 | | HIV-1 gp41 clade B double alanine mutant Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Gp41 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
