3E8D
 
 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, Glycogen synthase kinase-3 beta peptide, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4Y7L
 
 | | T6SS protein TssM C-terminal domain (869-1107) from EAEC | | Descriptor: | GLYCEROL, Type VI secretion protein IcmF, ZINC ION | | Authors: | Nguyen, V.S, Spinelli, S, Durand, E, Roussel, A, Cambillau, C. | | Deposit date: | 2015-02-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biogenesis and structure of a type VI secretion membrane core complex.
Nature, 523, 2015
|
|
8BWD
 
 | | Crystal structure of human Twisted gastrulation protein homolog 1 (TWSG1), crystal form 1 | | Descriptor: | SULFATE ION, Twisted gastrulation protein homolog 1 | | Authors: | Malinauskas, T, Rudolf, A.F, Moore, G, Eggington, H, Belnoue-Davis, H, El Omari, K, Woolley, R.E, Griffiths, S.C, Duman, R, Wagner, A, Leedham, S.J, Baldock, C, Ashe, H, Siebold, C. | | Deposit date: | 2022-12-06 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular mechanism of BMP signal control by Twisted gastrulation.
Nat Commun, 15, 2024
|
|
6GFX
 
 | | pVHL:EloB:EloC in complex with modified HIF-1a CODD peptide containing (3R,4S)-3-fluoro-4-hydroxyproline (ligand 13a) | | Descriptor: | Elongin-B, Elongin-C, FLUORINATED HYPOXIA-INDUCIBLE FACTOR 1 ALPHA PEPTIDE, ... | | Authors: | Castro, G.V, Testa, A, Ciulli, A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 3-Fluoro-4-hydroxyprolines: Synthesis, Conformational Analysis, and Stereoselective Recognition by the VHL E3 Ubiquitin Ligase for Targeted Protein Degradation.
J. Am. Chem. Soc., 140, 2018
|
|
8BWA
 
 | | Crystal structure of human Twisted gastrulation protein homolog 1 (TWSG1) in complex with platinum | | Descriptor: | PLATINUM (II) ION, Twisted gastrulation protein homolog 1 | | Authors: | Malinauskas, T, Rudolf, A.F, Moore, G, Eggington, H, Belnoue-Davis, H, El Omari, K, Woolley, R.E, Griffiths, S.C, Duman, R, Wagner, A, Leedham, S.J, Baldock, C, Ashe, H, Siebold, C. | | Deposit date: | 2022-12-06 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Molecular mechanism of BMP signal control by Twisted gastrulation.
Nat Commun, 15, 2024
|
|
5FIT
 
 | | FHIT-SUBSTRATE ANALOG | | Descriptor: | FRAGILE HISTIDINE TRIAD PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
5IK4
 
 | | Laminin A2LG45 C-form, Apo. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Briggs, D.C, Hohenester, E, Campbell, K.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural basis of laminin binding to the LARGE glycans on dystroglycan.
Nat.Chem.Biol., 12, 2016
|
|
6C2O
 
 | | Crystal structure of HCV NS3/4A protease variant Y56H in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, GLYCEROL, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.179 Å) | | Cite: | Clinical signature variant of HCV NS3/4A protease uses a novel mechanism to confer resistance
To be Published
|
|
4WWD
 
 | | High-resolution structure of the Co-bound form of the Y135F mutant of C. metallidurans CnrXs | | Descriptor: | CARBON DIOXIDE, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Volbeda, A, Coves, J, Maillard, A.P, Kuennemann, S, Grosse, C, Schleuder, G, Petit-Haertlein, I, de Rosny, E, Nies, D.H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding.
Metallomics, 7, 2015
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
5FFT
 
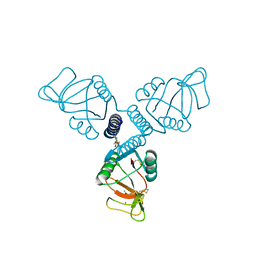 | | Crystal Structure of Surfactant Protein-A Y221A Mutant | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
4O5C
 
 | |
6A29
 
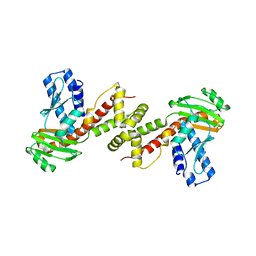 | | Crystal structure of PprA A139R mutant | | Descriptor: | DNA repair protein PprA | | Authors: | Adachi, M, Shibazaki, C, Shimizu, R, Arai, S, Satoh, K, Narumi, I, Kuroki, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans.
FASEB J., 33, 2019
|
|
5ESH
 
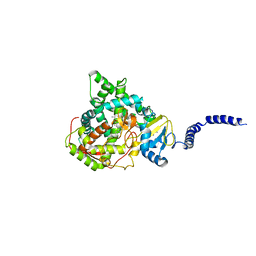 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) G73W mutant in complex with itraconazole | | Descriptor: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Impact of Homologous Resistance Mutations from Pathogenic Yeast on Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 62, 2018
|
|
5ESJ
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) G464S mutant complexed with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Impact of Homologous Resistance Mutations from Pathogenic Yeast on Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 62, 2018
|
|
5ESK
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) G464S mutant complexed with itraconazole | | Descriptor: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Impact of Homologous Resistance Mutations from Pathogenic Yeast on Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 62, 2018
|
|
6NKZ
 
 | |
6DNF
 
 | |
7PPF
 
 | | CRYSTAL STRUCTURE OF NAMPT IN COMPLEX WITH COMPOUND 8 | | Descriptor: | 1,2-ETHANEDIOL, N-[4-[(4R)-1,4-dimethyl-6-oxidanylidene-4,5-dihydropyridazin-3-yl]phenyl]-5,7-dihydropyrrolo[3,4-b]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Hillig, R.C. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A Novel NAMPT Inhibitor-Based Antibody-Drug Conjugate Payload Class for Cancer Therapy.
Bioconjug.Chem., 33, 2022
|
|
5I0V
 
 | |
465D
 
 | | STRUCTURE OF THE TOPOISOMERASE II POISON BOUND TO DNA | | Descriptor: | 9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P. | | Deposit date: | 1999-04-14 | | Release date: | 1999-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the topoisomerase II poison 9-amino-[N-(2-dimethylamino)ethyl]acridine-4-carboxamide bound to the DNA hexanucleotide d(CGTACG)2.
Biochemistry, 38, 1999
|
|
4M68
 
 | | Crystal structure of the mouse MLKL kinase-like domain | | Descriptor: | GLYCEROL, Mixed lineage kinase domain-like protein | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
1AAB
 
 | | NMR STRUCTURE OF RAT HMG1 HMGA FRAGMENT | | Descriptor: | HIGH MOBILITY GROUP PROTEIN | | Authors: | Hardman, C.H, Broadhurst, R.W, Raine, A.R.C, Grasser, K.D, Thomas, J.O, Laue, E.D. | | Deposit date: | 1995-10-28 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the A-domain of HMG1 and its interaction with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 34, 1995
|
|
6AKV
 
 | | Crystal structure of LysB4, the endolysin from Bacillus cereus-targeting bacteriophage B4 | | Descriptor: | CHLORIDE ION, LysB4, SULFATE ION, ... | | Authors: | Hong, S, Ha, N.-C. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of LysB4, an Endolysin fromBacillus cereus-Targeting Bacteriophage B4.
Mol. Cells, 42, 2019
|
|
1AFH
 
 | | LIPID TRANSFER PROTEIN FROM MAIZE SEEDLINGS, NMR, 15 STRUCTURES | | Descriptor: | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Gomar, J, Petit, M.C, Sodano, P, Sy, D, Marion, D, Kader, J.C, Vovelle, F, Ptak, M. | | Deposit date: | 1997-03-07 | | Release date: | 1997-05-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure and lipid binding of a nonspecific lipid transfer protein extracted from maize seeds.
Protein Sci., 5, 1996
|
|
