3GF9
 
 | | Crystal structure of human Intersectin 2 RhoGEF domain | | Descriptor: | Intersectin 2, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tong, Y, Tempel, W, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human Intersectin 2 RhoGEF domain
To be Published
|
|
4DTY
 
 | | cytochrome P450 BM3h-8C8 MRI sensor, no ligand | | Descriptor: | MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 8C8 | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
2O8V
 
 | | PAPS reductase in a covalent complex with thioredoxin C35A | | Descriptor: | Phosphoadenosine phosphosulfate reductase, Thioredoxin 1 | | Authors: | Chartron, J, Shiau, C, Stout, C.D, Carroll, K.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3'-Phosphoadenosine-5'-phosphosulfate Reductase in Complex with Thioredoxin: A Structural Snapshot in the Catalytic Cycle.
Biochemistry, 46, 2007
|
|
2OD9
 
 | | Structural Basis for Nicotinamide Inhibition and Base Exchange in Sir2 Enzymes | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, H4 peptide, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R, Sanders, B.D. | | Deposit date: | 2006-12-21 | | Release date: | 2007-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
2O95
 
 | | Crystal Structure of the Metal-Free Dimeric Human Mov34 MPN domain (residues 1-186) | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 26S proteasome non-ATPase regulatory subunit 7, DODECAETHYLENE GLYCOL, ... | | Authors: | Sanches, M, Alves, B.S.C, Zanchin, N.I.T, Guimaraes, B.G. | | Deposit date: | 2006-12-13 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of the Human Mov34 MPN Domain Reveals a Metal-free Dimer
J.Mol.Biol., 370, 2007
|
|
4DJX
 
 | |
4H95
 
 | |
3R96
 
 | |
3GAC
 
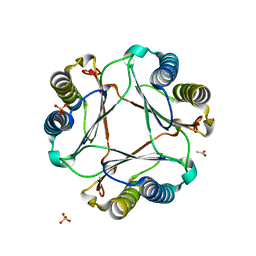 | | Structure of mif with HPP | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ACETIC ACID, Macrophage migration inhibitory factor-like protein, ... | | Authors: | Zhou, Y.-F, Su, X.-D, Shao, D, Wang, H. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host
Mol.Immunol., 47, 2010
|
|
3RGF
 
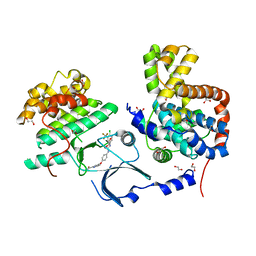 | | Crystal Structure of human CDK8/CycC | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Blaesse, M, Huber, R, Maskos, K. | | Deposit date: | 2011-04-08 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of CDK8/CycC Implicates Specificity in the CDK/Cyclin Family and Reveals Interaction with a Deep Pocket Binder.
J.Mol.Biol., 412, 2011
|
|
3R9I
 
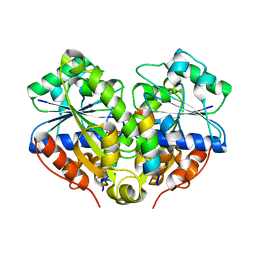 | | 2.6A resolution structure of MinD complexed with MinE (12-31) peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
4H9E
 
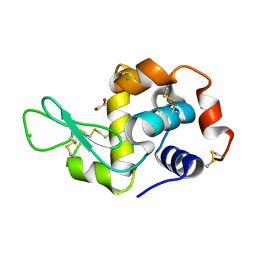 | | Radiation damage study of lysozyme - 0.84 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1998 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4DLC
 
 | | Crystal Structure of Trypanosoma brucei dUTPase with dUMP, MgF3- transition state analogue, and Mg2+ | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION, ... | | Authors: | Hemsworth, G.R, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2012-02-06 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | On the catalytic mechanism of dimeric dUTPases.
Biochem.J., 456, 2013
|
|
3RAV
 
 | | Horse spleen apo-ferritin with bound Pentobarbital | | Descriptor: | 5-ethyl-5-[(2S)-pentan-2-yl]pyrimidine-2,4,6(1H,3H,5H)-trione, CADMIUM ION, Ferritin light chain, ... | | Authors: | Oakley, S.H, Vedula, L.S, Xi, J, Liu, R, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution view of barbiturate recognition by a protein binding site
to be published
|
|
3RHH
 
 | | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP
To be Published
|
|
4DO5
 
 | | Pharmacological chaperones for human alpha-N-acetylgalactosaminidase | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clark, N.E, Garman, S.C. | | Deposit date: | 2012-02-09 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Pharmacological chaperones for human alpha-N-acetylgalactosaminidase
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RIE
 
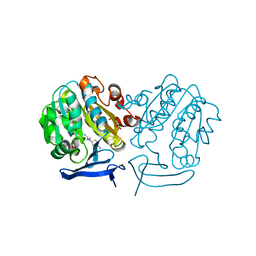 | | The structure of Plasmodium falciparum spermidine synthase in complex with 5'-methylthioadenosine and N-(3-aminopropyl)-trans-cyclohexane-1,4-diamine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, ... | | Authors: | Williams, M, Sprenger, J, Persson, L, Louw, A.I, Birkholtz, L, Al-Karadaghi, S. | | Deposit date: | 2011-04-13 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel inhibitor of Plasmodium falciparum spermidine synthase: a twist in the tail.
Malar J, 14, 2015
|
|
4DWH
 
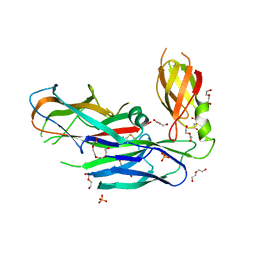 | | Structure of the major type 1 pilus subunit FIMA bound to the FIMC (2.5 A resolution) | | Descriptor: | Chaperone protein fimC, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Scharer, M.A, Puorger, C, Crespo, M, Glockshuber, R, Capitani, G. | | Deposit date: | 2012-02-24 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quality control of disulfide bond formation in pilus subunits by the chaperone FimC.
Nat.Chem.Biol., 8, 2012
|
|
3GFW
 
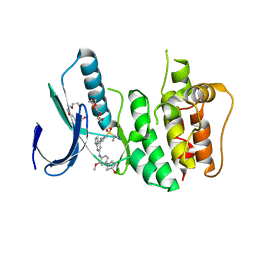 | | Crystal Structure of Human Dual Specificity Protein Kinase (TTK) in complex with a pyrolo-pyridin ligand | | Descriptor: | 1-(4-(4-(2-(isopropylsulfonyl)phenylamino)-1H-pyrrolo[2,3-b]pyridin-6-ylamino)-3-methoxyphenyl)piperidin-4-ol, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Filippakopoulos, P, Soundararajan, M, Choi, H, Keates, T, Elkins, J.M, King, O, Fedorov, O, Picaud, S.S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Grey, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function.
Nat.Chem.Biol., 6, 2010
|
|
2OKA
 
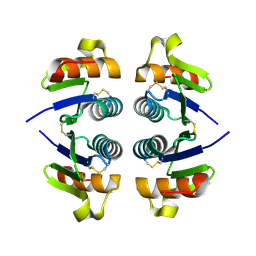 | | Crystal structure of Q9HYQ7_PSEAE from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium target PaR82 | | Descriptor: | Hypothetical protein | | Authors: | Benach, J, Neely, H, Seetharaman, J, Chen, X.C, Fang, Y, Cunningham, K, Owens, L, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-01-16 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Q9HYQ7_PSEAE from Pseudomonas aeruginosa
To be Published
|
|
4DQG
 
 | |
3G8D
 
 | | Crystal structure of the biotin carboxylase subunit, E296A mutant, of acetyl-COA carboxylase from Escherichia coli | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, MAGNESIUM ION, ... | | Authors: | Chou, C.Y, Yu, L.P, Tong, L. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
J.Biol.Chem., 284, 2009
|
|
2OGR
 
 | |
4DQP
 
 | | Ternary complex of Bacillus DNA Polymerase I Large Fragment, DNA duplex, and ddCTP (paired with dG of template) | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*C*AP*TP*GP*GP*GP*AP*GP*TP*CP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DOC))-3'), ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-02-16 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides.
J.Biol.Chem., 287, 2012
|
|
3RFW
 
 | | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally-related SurA-like chaperones in the human pathogen Campylobacter jejuni | | Descriptor: | Cell-binding factor 2 | | Authors: | Kale, A, Phansopa, C, Suwannachart, C, Craven, C.J, Rafferty, J, Kelly, D.J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The virulence factor PEB4 and the periplasmic protein Cj1289 are two structurally-related SurA-like chaperones in the human pathogen Campylobacter jejuni
To be Published
|
|
