2QWS
 
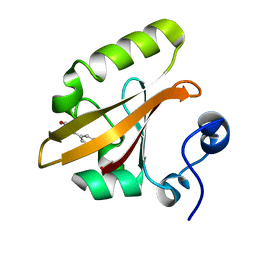 | |
2E4M
 
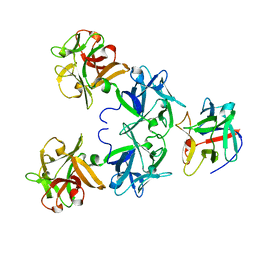 | | Crystal structure of hemagglutinin subcomponent complex (HA-33/HA-17) from Clostridium botulinum serotype D strain 4947 | | Descriptor: | HA-17, Main hemagglutinin component | | Authors: | Hasegawa, K, Watanabe, T, Suzuki, T, Yamano, A, Niwa, K, Ohyama, T. | | Deposit date: | 2006-12-13 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Subunit Structure of Clostridium botulinum Serotype D Toxin Complex with Three Extended Arms
J.Biol.Chem., 282, 2007
|
|
2FO3
 
 | | Plasmodium vivax ubiquitin conjugating enzyme E2 | | Descriptor: | Ubiquitin-conjugating enzyme | | Authors: | Dong, A, Zhao, Y, Lew, J, Kozieradski, I, Alam, Z, Melone, M, Wasney, G, Vedadi, M, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Bochkarev, A, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
2FW2
 
 | | Catalytic domain of CDY | | Descriptor: | Testis-specific chromodomain protein Y 2 | | Authors: | Min, J.R, Antoshenko, T, Wu, H, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of catalytic domain of Human Testis-specific chromodomain protein Y.
To be Published
|
|
2FYT
 
 | | Human HMT1 hnRNP methyltransferase-like 3 (S. cerevisiae) protein | | Descriptor: | Protein arginine N-methyltransferase 3, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Min, J.R, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-08 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human HMT1 hnRNP
methyltransferase-like 3 in complex with SAH.
To be Published
|
|
4K3Z
 
 | |
6QG5
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model C) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QKN
 
 | | Structure of the azide-inhibited form of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae | | Descriptor: | AZIDE ION, CALCIUM ION, Cytochrome-c peroxidase, ... | | Authors: | Carvalho, A.L, Romao, M.J, Pauleta, S, Nobrega, C. | | Deposit date: | 2019-01-29 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mixed-valence, active form, of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae
To Be Published
|
|
6QKS
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Apo | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6QLA
 
 | | CRYSTAL STRUCTURE OF THE PMGL2 ESTERASE (point mutant 1) FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Boyko, K.M, Garsia, D, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of PMGL2 esterase from the hormone-sensitive lipase family with GCSAG motif around the catalytic serine.
Plos One, 15, 2020
|
|
6Q9T
 
 | | Protein-aromatic foldamer complex crystal structure | | Descriptor: | Aromatic foldamer, Carbonic anhydrase 2, ZINC ION | | Authors: | Post, S, Langlois d'Estaintot, B, Fischer, L, Granier, T, Huc, I. | | Deposit date: | 2018-12-18 | | Release date: | 2019-09-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure Elucidation of Helical Aromatic Foldamer-Protein Complexes with Large Contact Surface Areas.
Chemistry, 25, 2019
|
|
6QMQ
 
 | | NF-YB/C Heterodimer in Complex with NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | Nuclear transcription factor Y subunit alpha, Nuclear transcription factor Y subunit beta, Nuclear transcription factor Y subunit gamma, ... | | Authors: | Kiehstaller, S, Jeganathan, S, Pearce, N.M, Wendt, M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2019-02-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Constrained Peptides with Fine-Tuned Flexibility Inhibit NF-Y Transcription Factor Assembly.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QQ8
 
 | |
8H28
 
 | | Crystal structure of the K87V mutant of cytochrome c' from Shewanella benthica DB6705 | | Descriptor: | Class II cytochrome c, HEME C | | Authors: | Fujii, S, Sakaguchi, R, Oki, H, Kawahara, K, Ohkubo, T, Fujiyoshi, S, Sambongi, Y. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Contribution of a surface salt bridge to the protein stability of deep-sea Shewanella benthica cytochrome c'.
J.Struct.Biol., 215, 2023
|
|
6QU0
 
 | | Structure of azoreductase from Bacillus sp. A01 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase, GLYCEROL, ... | | Authors: | Savino, S, Fraaije, M.W. | | Deposit date: | 2019-02-26 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic and Crystallographic Studies of Azoreductase AzoA fromBacillus wakoensisA01.
Acs Chem.Biol., 15, 2020
|
|
6QWB
 
 | | Crystal structure of KPC-4 complexed with relebactam (16 hour soak) | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2~{S})-5-azanylidene-2-(piperidin-4-ylcarbamoyl)piperidine-1-carboxylic acid, Beta-lactamase, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Molecular Basis of Class A beta-Lactamase Inhibition by Relebactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
6QWD
 
 | | Crystal structure of KPC-3 | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular Basis of Class A beta-Lactamase Inhibition by Relebactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
6QM8
 
 | | Leishmania tarentolae proteasome 20S subunit apo structure | | Descriptor: | Proteasome alpha1 chain, Proteasome alpha2 chain, Proteasome alpha3 chain, ... | | Authors: | Rowland, P, Goswami, P. | | Deposit date: | 2019-02-01 | | Release date: | 2019-04-17 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Preclinical candidate for the treatment of visceral leishmaniasis that acts through proteasome inhibition.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8HKD
 
 | |
6QUG
 
 | | GHK tagged MBP-Nup98(1-29) | | Descriptor: | COPPER (II) ION, Maltodextrin-binding protein,Nucleoporin, putative, ... | | Authors: | Huyton, T, Gorlich, D. | | Deposit date: | 2019-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The copper(II)-binding tripeptide GHK, a valuable crystallization and phasing tag for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6QMP
 
 | | NF-YB/C Heterodimer in Complex with NF-YA Peptide | | Descriptor: | Nuclear transcription factor Y subunit alpha, Nuclear transcription factor Y subunit beta, Nuclear transcription factor Y subunit gamma | | Authors: | Kiehstaller, S, Jeganathan, S, Pearce, N.M, Wendt, M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2019-02-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Constrained Peptides with Fine-Tuned Flexibility Inhibit NF-Y Transcription Factor Assembly.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4XJE
 
 | |
3N0H
 
 | | hDHFR double mutant Q35S/N64F Trimethoprim Binary Complex | | Descriptor: | Dihydrofolate reductase, SULFATE ION, TRIMETHOPRIM | | Authors: | Cody, V. | | Deposit date: | 2010-05-14 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystallographic analysis reveals a novel second binding site for trimethoprim in active site double mutants of human dihydrofolate reductase.
J.Struct.Biol., 176, 2011
|
|
6QPN
 
 | | Adenovirus species D serotype 49 Fiber-Knob | | Descriptor: | Fiber, SULFATE ION | | Authors: | Baker, A.T, Rizkallah, P.J. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The Fiber Knob Protein of Human Adenovirus Type 49 Mediates
Highly Efficient and Promiscuous Infection of Cancer Cell Lines
Using a Novel Cell Entry Mechanism
Journal of Virology, 95, 2021
|
|
8HM5
 
 | |
